Figure 2.
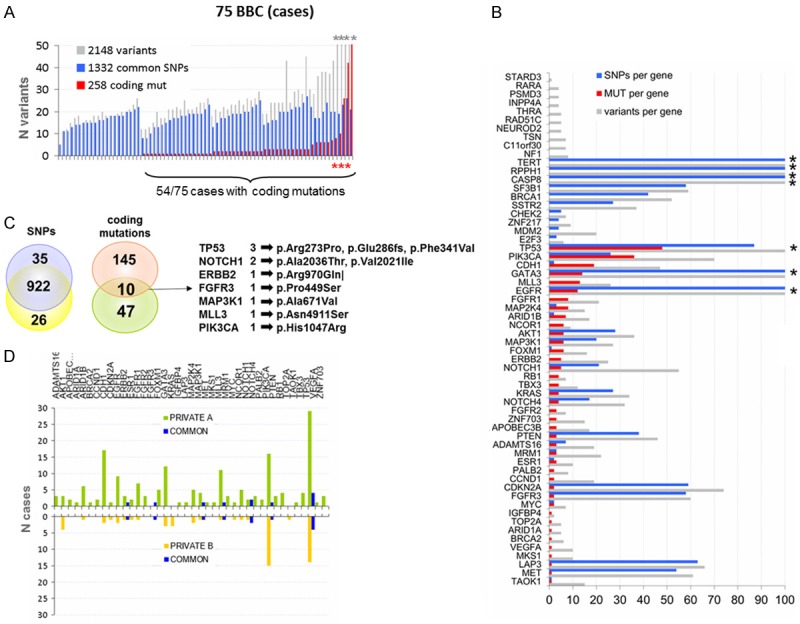
Distribution of NGS variants in BBC. A. Distribution of variants of any type in the study cases. All variants, SNPs and coding mutations are shown per case. For similar sequencing efficiency metrics (total reads, on target reads, uniformity of reads), 5-214 variants were identified per case (mean ± SD: 23 ± 30; median: 22), up to 54 in the same gene (mean ± SD: 15 ± 7; median: 12). Asterisks: truncated Y-axis for cases with >50 variants. B. Distribution of variants per gene. Grey bars: total number of variants of any type. Coding mutations were identified in 41 out of 61 genes in the MPS panel (red bars). The total number of mutations per gene varied from 1 to 49 (median: 3 mutations per gene). In additional 10 genes, only SNPs were present (blue bars). The number of SNPs per sample ranged from 10-20. Asterisks: >100 variants for these genes. C. Common and private SNPs and coding mutations. SNPs were preserved at 94% in both sides (922/983 comparable SNPs); by contrast, only 4.9% common coding mutations were observed (10/202 comparable mutations). The remaining 192 comparable mutations were present unilaterally, as shown. D. Distribution of gene coding mutations on either side; common, private A and private B mutations represented by blue, green and orange bars, respectively.
