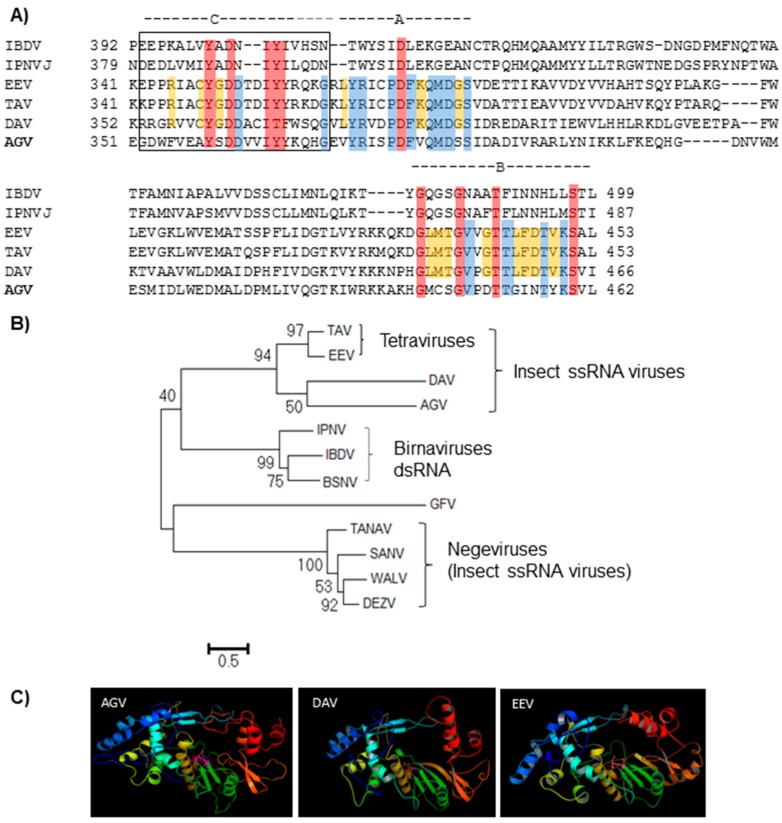
Figure 2.
ApGlV2 has a non-canonical (C-A-B) RdRp, similar to those of some other insect viruses. (A) The conservation of the amino acid residues between ApGlV2, the tetraviruses; Thosea asigna virus (TAV) and Euprosterna elaeasa virus (EEV); Drosophila A virus (DAV) and birnaviruses; Infectious pancreatic necrosis virus (IPNV) and Infectious bursal disease virus (IBDV) are highlighted. Yellow highlights show conservation between EEV, TAV, and DAV but not ApGlV2 while blue highlights show conservation of EEV, TAV, and DAV with ApGlV2. Red highlights show conservation of residues between all viruses. All highlighted amino acids are key RdRp motifs. (B) Phylogenetic tree of ApGlV2 RdRp with closely related viruses and other viruses with permuted RdRp based on BLAST results of amino acid identity. Phylogenetic trees were constructed using MEGA 6.06 with the maximum likelihood method. Thosea asigna virus (TAV, accession AAQ14329.1) and EEV (AF461742_1) are closely related to but phylogenetically distinct from DAV (YP003038595.1) and ApGlV2 (AGV). The RdRps of these insect viruses are clustered with Birnaviruses, e.g., Infectious pancreatic necrosis virus (IPNV; AAV48847.1), Infectious bursal disease virus (IBDV; ACS44343.1), and Blotched snakehead virus (BSNV; YP052864). In contrast, the RdRps of negeviruses with permuted RdRps, Dezidougou virus (DEZV, AFI24669.1), Santana virus (SANV, AFI24675.1), Wallerfield virus (WALV, AIS40860.1), and Tanay virus (TANNV, YP_009028558.1) are close to a plant virus, Grapevine fleck virus (GFV, CAC84400.1), forming a distinct branch and indicating that the RdRps of negeviruses are distinct from that of ApGlV2; (C) The predicted RdRp structures for ApGlV2 (AGV), DAV, and EEV are similar. Homology modeling of tertiary protein structures of RdRp from ApGlV2, Drosophila A virus (DAV), and Euprosterna elaeasa virus (EEV). Images were generated using the LOMETS server.