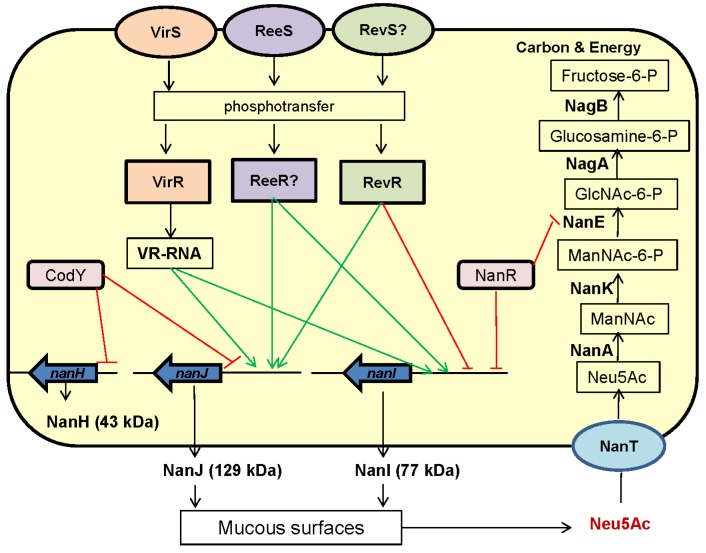
Figure 2.
A proposed model for regulation of expression of sialidase genes and the pathway for sialic acid metabolism in C. perfringens. Exosialidases generate free sialic acid from mucus or host cell surfaces [33,40]. The free sialic acid is then transported into C. perfringens where it is metabolized to fructose-6-P [40]. There is evidence that VirS/VirR, RevR, ReeS, NanR, and CodY systems directly or indirectly affect sialidase production, although inter-relationships between these regulators are unclear [40,41,43,44,45,46]. The VirS/VirR two component system acts as a positive regulator of the vrr gene, which encodes VR-RNA. VR-RNA is then a positive regulator of nanI and nanJ expression [41,42]. The ReeS sensor kinase positively regulates nanI and nanJ gene expression, presumably by a putative transcriptional regulator named ReeR [43]. RevS positively regulates nanJ expression but negatively regulates nanI expression [44]. CodY represses nanH and nanJ expression [45]. Based on sequence homology comparisons with similar regulators in other bacteria, NanR may repress nanI expression and the sialic acid metabolism pathway, but there is no direct experimental evidence yet to support this hypothesis. Green lines indicate positive regulation while red lines indicate negative regulation.