Figure 1. Key Figure, Strategy for zebrafish genome engineering with CRISPR-Cas9.
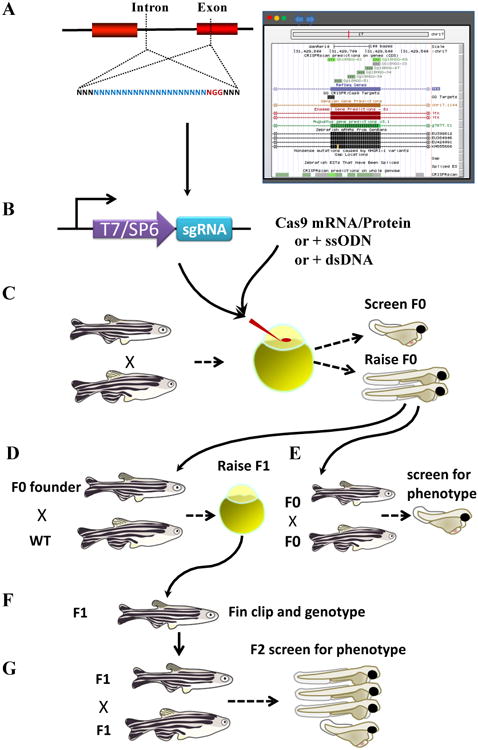
Schematic depiction of the steps in a zebrafish CRISPR experiment. The target is selected (A), the sgRNA is synthesized by in vitro RNA synthesis (B), and injected together with Cas9 protein or cas9 mRNA into the 1-cell stage zebrafish (C). Mutagenesis efficiency is evaluated for each sgRNA using various methods including T7 endonuclease, heteroduplex mobility shift (HMA), and sequencing (D). The phenotype of the F0 animals can also be assessed. Successfully mutagenized F0 animals are raised to adulthood and crossed to WT animals; or crossed to other F0 and the phenotype of the offspring assessed (E). F1 animals are genotyped and sequenced (F) and crossed either to siblings or to independently generated alleles in the same gene (G).
