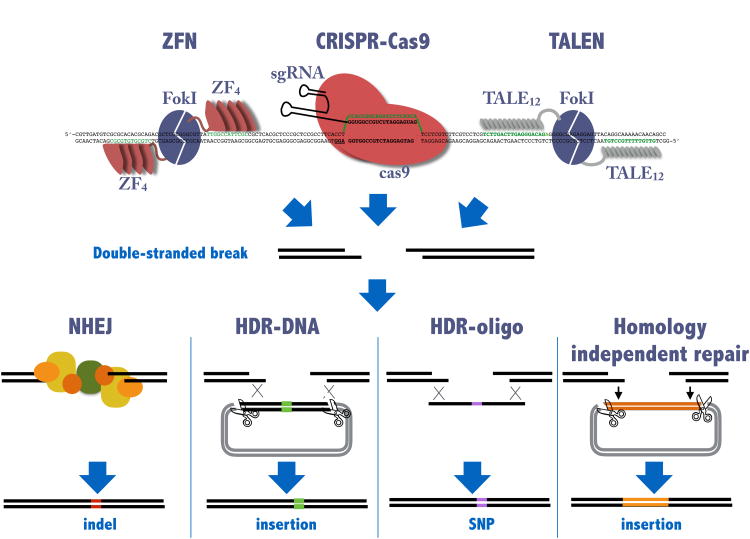
Figure I. Genomic engineering using ZFN, TALEN, and the CRISPR-Cas9 endonucleases.
The three different endonucleases can be designed to site-specifically recognize a single-site in a vertebrate genome. Both ZFN and TALEN are constructed as dimers recognizing two sites in a head-to-head orientation (indicated in green). Modular ZF or TALE domains are used to recognize the specific locus while the FokI endonuclease domain cuts the target. The FokI dimerization interface has been engineered to only allow formation of heterodimers. Expression of both proteins is required to make a functional endonuclease. CRISPR-Cas9 is a complex between the Cas9 protein and a small RNA molecule, the sgRNA. Only the 5′ end of the sgRNA need be changed to change the specificity of this endonuclease. All three endonucleases generate double stranded DNA breaks (DSB) which are repaired by the endogenous DNA repair machinery. This can occur without a template by NHEJ mechanisms or can be templated by exogenous DNA for example in HDR or can incorporate exogenously supplied DNA in a homology independent process. The exact mechanisms of repair are currently being investigated.