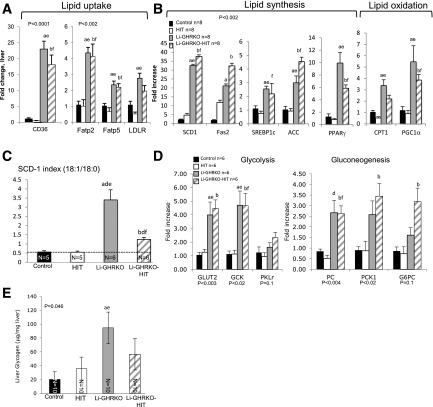
Figure 5.
Hepatic Igf-1 transgene does not resolve hepatic lipid metabolism in Li-GHRKO mice. A and B: Liver gene expression of key players in lipid metabolism was determined by real-time PCR (n = 8/genotype). C: SCD-1 index determined by the ratio of hepatic stearic and oleic acid concentrations determined by GC/MS. D: Hepatic gene expression of key players in glycolysis (GLUT2, glucokinase [GCK], and liver pyruvate kinase [PKLr]) and gluconeogenesis (PC, PCK1, and glucose 6 phosphatase [G6PC]). Liver RNA extracted from 16- to 24-week-old male mice and processed for real-time PCR assay. E: Hepatic glycogen content assayed in livers of 16-week-old male mice. Data presented as mean ± SEM. N indicates sample size. Significance accepted at P < 0.05: control vs. Li-GHRKO (a), control vs. Li-GHRKO-HIT (b), Li-GHRKO vs. Li-GHRKO-HIT (d), Li-GHRKO vs. HIT (e), and Li-GHRKO-HIT vs. HIT (f). ACC, acetyl-CoA carboxylase; Fas2, FA synthase 2; Fatp, FA transport protein; LDLR, LDL receptor; PGC1α, peroxisome proliferator–activated receptor-γ coactivator 1α; PPARγ, peroxisome proliferator–activated receptor γ; SREBP1c, sterol regulatory element–binding protein 1c.