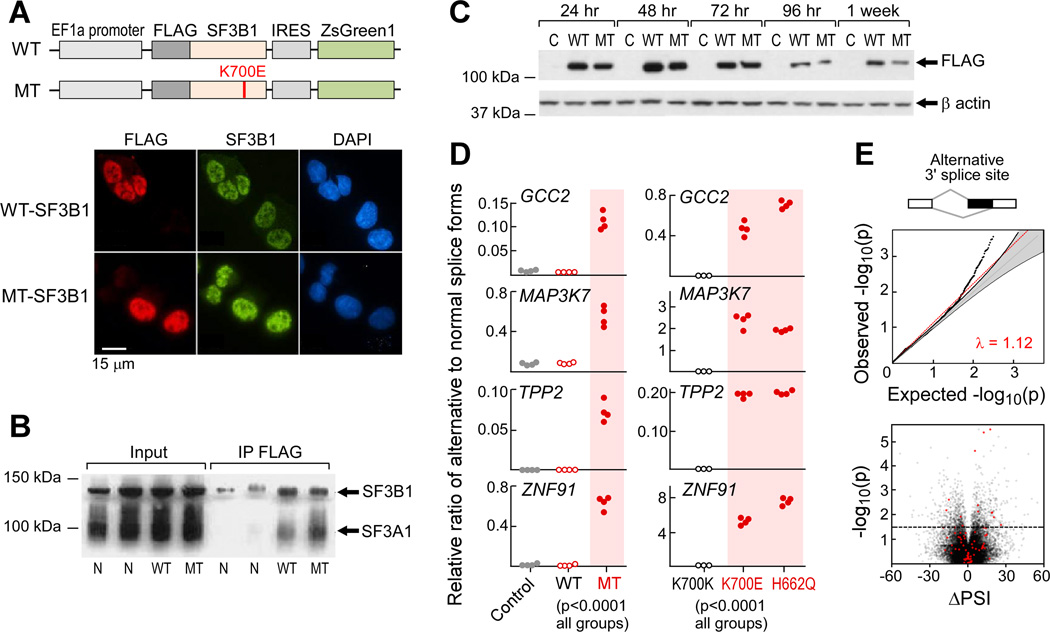
Figure 2. Expression of mutant SF3B1 causes alternative splicing.
(A) Schematics of wild-type (WT) and K700E mutated (MT) SF3B1 expression constructs (top) and immunoflourescence staining of FLAG-tagged SF3B1 in K562 cells that were nucleofected with SF3B1 constructs (bottom).
(B) Cell lysate from HeLa cells overexpressing either WT or MT SF3B1 was immunprecipated with anti-FLAG antibody and probed with anti-SF3B1 antibody.
(C) FLAG-tagged mutant and wild-type SF3B1 protein were transiently expressed in HEK293T cells and detected by immunoblotting.
(D) Expression of alternative splicing associated with SF3B1 mutations in transfected K562 cells (left, n=4 for each group) and in isogenic Nalm-6 cells (right) was assessed with quantitative RT-PCR assays.
(E) Analysis of bulk poly-A selected RNA-seq of K562 cells expressing SF3B1-K700E. Top panel- Q–Q plot of alternative 3’ splice sites between empirical p value of observed and expected spliced events. Lower panel - volcano plot of ΔPSI in relation to significance between wild-type and mutant SF3B1 overexpression in K562 cells for each splicing events. Red dots - significantly differentially spliced events also identified in the primary CLL analysis.
See also Figure S2.