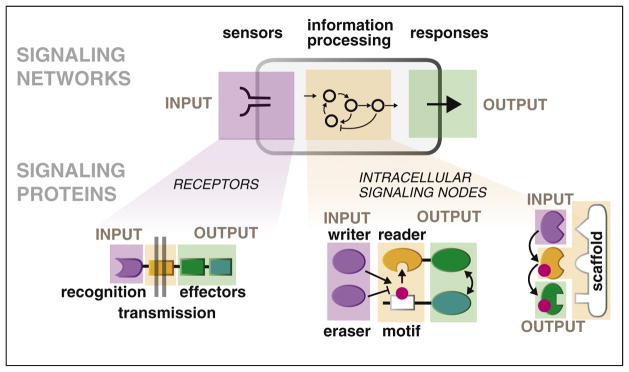
Figure 1.
Hierarchical organization of signaling systems: cells and individual proteins as input/output nodes. At any scale, a signaling system must have three components — it must have sensors to receive INPUT, an information-processing layer that decides what to make of this information, and an OUTPUT function. These components are found in individual signaling molecules, which detect and effect particular upstream and downstream molecular partners. In receptors that span the cell’s membrane, ligand binding to extracellular domains (INPUT) rapidly regulates the activity of intracellular effector domains (OUTPUT). Similarly, posttranslationally-regulated binding motifs link the activities of upstream enzymes that ‘write’ and ‘erase’ the posttranslational mark (INPUT; e.g. kinases and phosphatases) to recruitment of dedicated ‘reader’ domains (OUTPUT). The same classes of components are found in signaling networks and whole cells, but in this case receptor molecules function as INPUT sensors, networks of intracellular proteins function as the information processing layer, and various cellular response modules control OUTPUT.