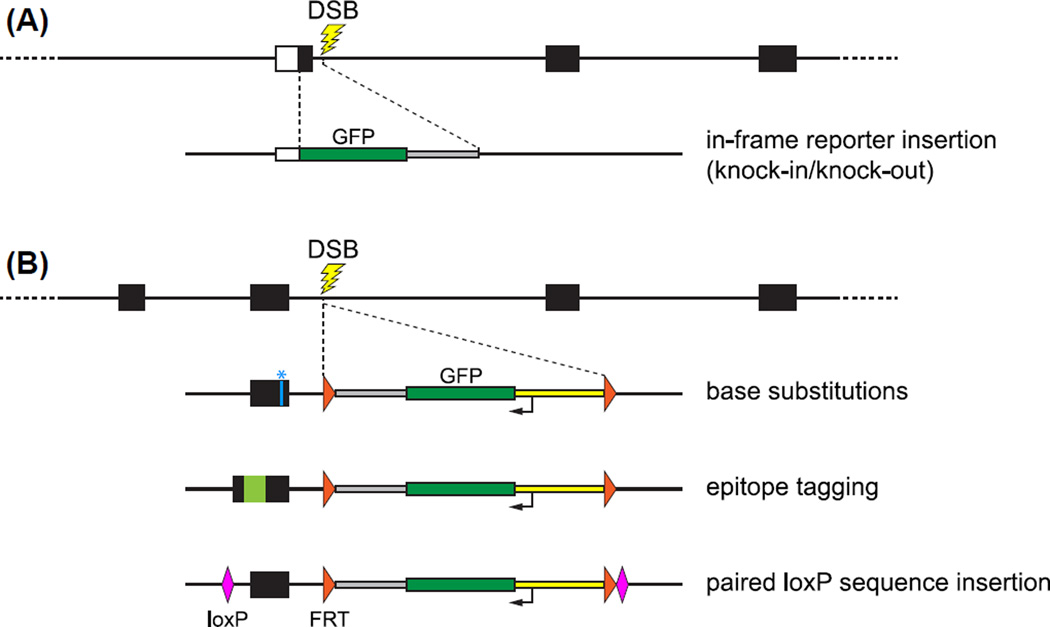
FIGURE 2. Examples of genome modifications produced by DSB-stimulated homologous recombination (HR) using dsDNA as donor template molecules.
Recombination events are stimulated by targeted cleavage (lightning bolts) of the genome with programmable nucleases. The top line of each panel represents the host genome, with boxes indicating exons (white, 5′ untranslated region; black, coding sequences). Below the genome cartoons are examples of dsDNA donor templates that have been used to generate the indicated types of edited alleles. Except as indicated, donor sequences are identical to those of the genome: green boxes indicate GFP coding sequences, gray boxes indicate transcription termination sequences, yellow boxes indicate α-crystallin promoter sequences, orange arrowheads are FRT sites, and pink diamonds are loxP sites. (A) Generation of a knockin/knockout allele. The DSB was induced in intron sequences just 3′ to the first exon. Recombination with donor sequences introduced GFP reporter sequences in-frame just downstream of the endogenous translation initiation codon. Animals inheriting this kind of edited allele can be recognized by virtue of expression of GFP under control of the endogenous promoter. (B) Use of a linked reporter gene to tag genomic modifications. In addition to the desired editing events, donor DNAs can be designed so they introduce into the host genome a small reporter gene (here α-crystallin::GFP flanked by directly repeated FRT sites). Edited alleles that incorporate precise alteration of coding sequences (blue *–base substitutions), in-frame introduction of epitope sequences (pale green–epitope tagging), or introduction of a pair of loxP recombination sites flanking an exon can be identified by expression of the linked tissue–specific reporter gene. Screening for GFP fluorescence allows for the efficient recovery of phenotypically silent modification events and obviates the need for laborious DNA-based screening of F1 individuals. Following identification of successful editing events, the reporter gene can be excised using FLP-mediated recombination. (See color plate)