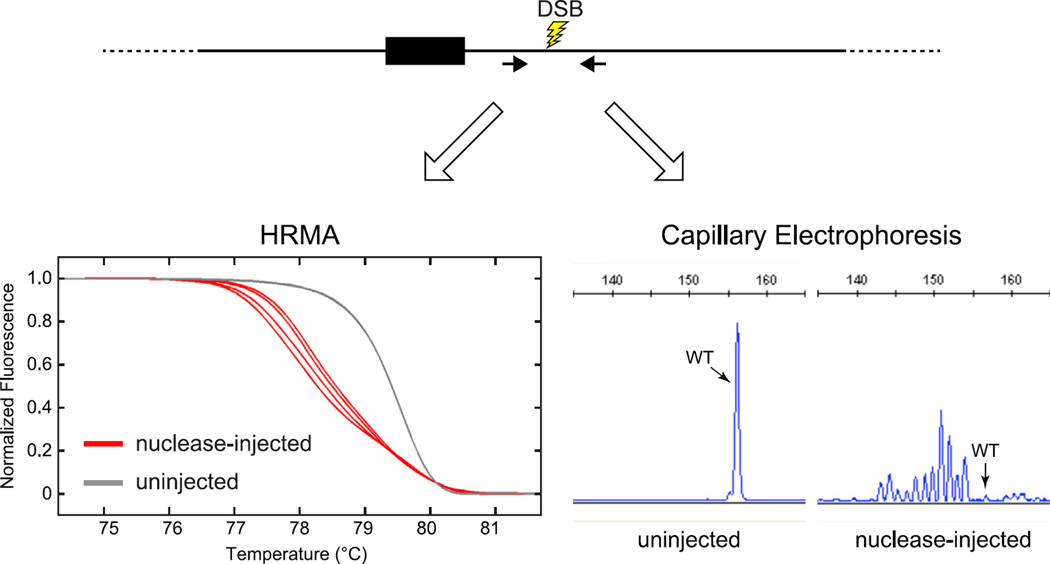
FIGURE 4. PCR-based methods to detect DSB repair-induced mutations.
Prior to conducting a genome-editing experiment, the in vivo activity of the programmable nuclease used to induce DSBs is assayed by the induction of small indels at the nuclease target site. Several methods are available for detecting repair-induced mutations. We use HRMA (Dahlem et al., 2012) or capillary electrophoresis (Carrington et al., 2015) for this purpose. Genomic DNA is isolated from individual 1 dpf embryos that had been injected at the one-cell stage with nuclease. PCR primers (black arrows) are used to amplify a 90–150-bp product that is centered on the nuclease target site. DNA from individual uninjected or nuclease-injected embryos is amplified in the presence of a dsDNA-binding dye (HRMA) or with fluorescently labeled primers (capillary electrophoresis). For HRMA, PCR products are denatured and renatured, and duplexes with mismatches are detected by their altered thermal denaturation profile (red curves (black in print versions)). For capillary electrophoresis analysis, products are resolved according to size, and the fraction of amplicons with altered size can be determined. Representative HRMA and capillary electrophoresis traces indicate a highly active nuclease.