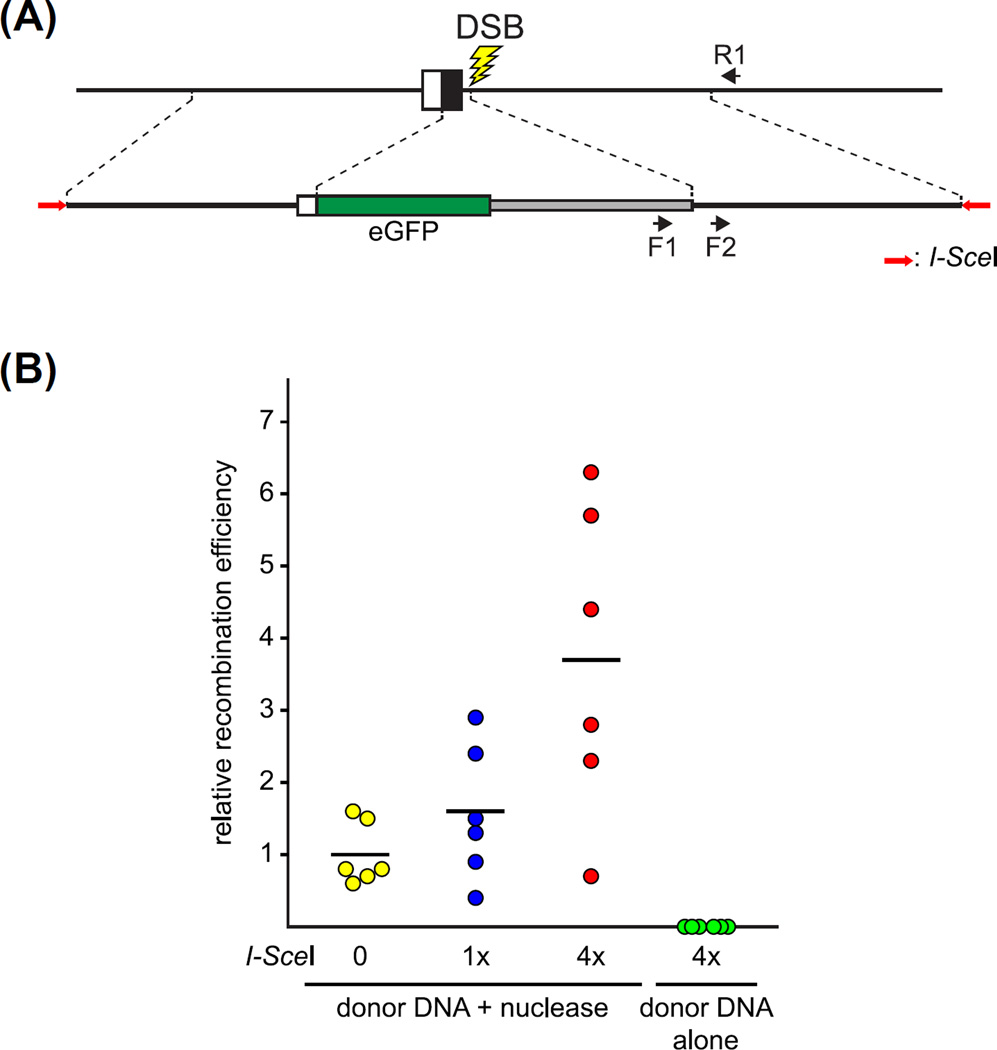
FIGURE 5. Enhancement of genome editing using donor molecules predigested with I-SceI meganuclease.
(A) Schematic representation of a genome-editing event to produce a reporter knockin/knockout allele, as in Fig. 2A. Donor DNA sequences were flanked by a pair of head-to-head oriented I-SceI sites (red arrows) present within the pKHR4 plasmid vector (Fig. 6). The relative abundance of edited alleles within the genomes of injected F0 embryos was determined following quantitative PCR (qPCR). Primers used for qPCR are depicted as black arrows: the F1/R1 pair specifically amplifies the edited allele, whereas the F2/R1 pair amplifies edited as well as unedited forms of the endogenous locus. (B) Genomic editing is enhanced by I-SceI digestion of a donor plasmid prior to injection. The donor plasmid was digested with increasing amounts of I-SceI enzyme in vitro and subsequently injected with programmable nuclease into zygotes. As a control, I-SceI-digested donor plasmid was injected alone, without added nuclease to target the host genome. The fraction of edited alleles (detected with the F1/R1 primer pair) relative to total targeted loci (detected with the F2/R1 primer pair) present in injected 2 dpf embryos was determined by qPCR. The relative recombination efficiency was determined by normalizing the mean fraction of edited alleles following injection of nuclease and undigested donor plasmid DNA to 1.0. For each condition, six individual embryos were analyzed (circles) and the mean relative recombination efficiency is indicated (horizontal dash). Unpaired t-test analysis indicated that digestion of donor DNA with 4× enzyme significantly stimulated the production of edited alleles as compared with untreated donor DNA (p < 0.01). Digestion with 1× enzyme did not yield a significant increase in recombination efficiency as compared with untreated DNA. Injection of I-SceI-digested DNA without programmable nuclease failed to produce a detectable level of edited target alleles. (See color plate)