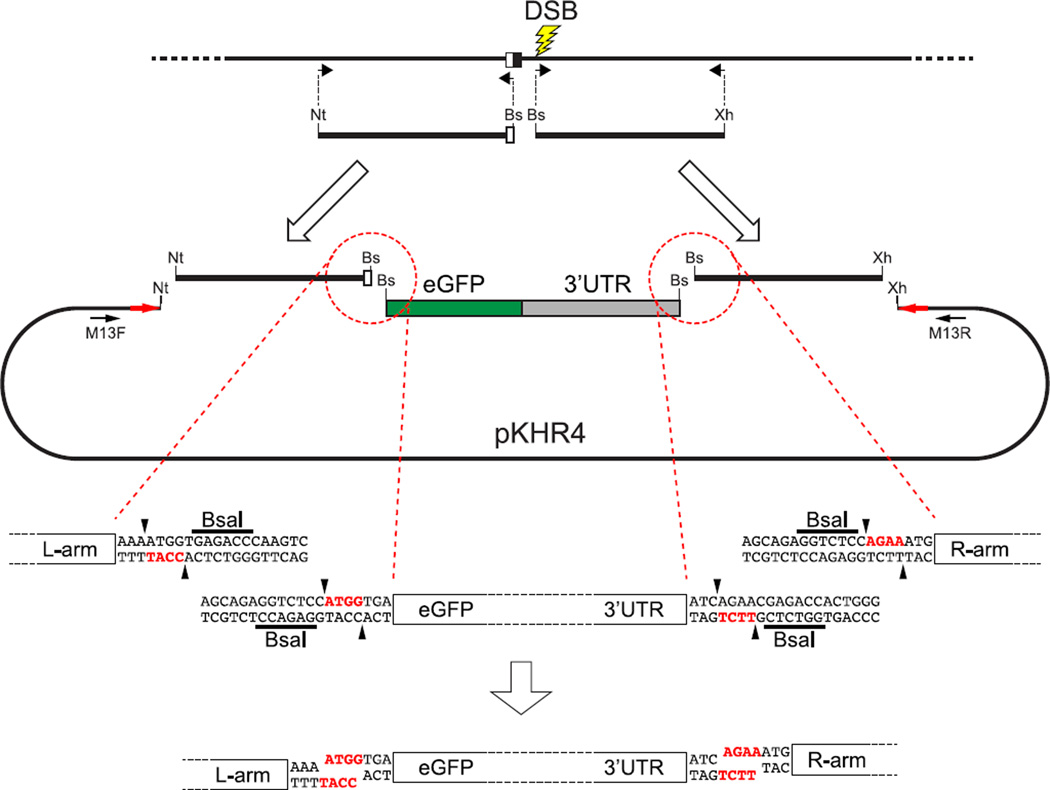
FIGURE 7. Preparation of a donor plasmid for targeted reporter integration.
To integrate a reporter gene consisting of eGFP coding sequences (green (dark gray in print versions)) and translation/transcription termination sequences (gray) at a specific target site, about 1 kbp of genomic sequences upstream and downstream of the reporter integration site are prepared by PCR amplification for left and right homology arms, respectively. The homology arms should include a mutated nuclease target sequence, so that integration of donor sequences will produce an edited allele that cannot be cleaved by the nuclease. Each amplified fragment is bordered by unique restriction enzyme recognition sequences derived from PCR primers. In this case, the left-arm fragment is bordered by NotI (Nt) and BsaI (Bs) sites, and the right arm has BsaI and XhoI (Xh) sites at its ends. BsaI is a type-II restriction enzyme that produces a staggered cut next to the enzyme recognition site. As a result, it can be used to generate unique, complementary protruding ends (red letters (black in print versions)) independent of the enzyme recognition sequence (underlined). The reporter sequence is prepared as a middle fragment by PCR amplification with ends containing BsaI recognition sites whose digestion would yield protruding ends complementary to the digested left- and right-arm fragments. Restriction enzyme-digested amplicons are individually purified. Because each digested end is complementary to a unique partner fragment, ligation of the digested fragments leads to ordered assembly, and a single cloning step is used to assemble the homology arms and the reporter middle fragment in correct sequence into the pKHR4 backbone, which had been predigested with NotI and XhoI.