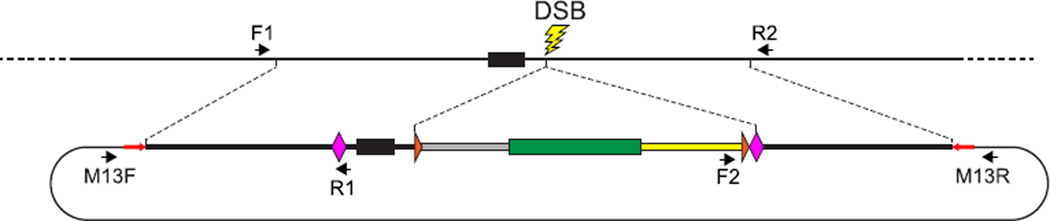
FIGURE 9. Diagnostic amplification to identify a precisely edited allele.
Once a candidate edited allele has been detected either by expression of the linked reporter gene or by preliminary genomic analysis, detailed PCR and sequence analysis of the targeted locus is performed to confirm that a precisely edited allele has been generated. Recombination events that correctly modify the targeted locus can be detected with primer pairs such as F1/R1 and F2/R2, which specifically amplify host–donor junction fragments and are expected to be present only in genomes harboring a precisely edited allele. In addition, to detect imprecise integration events, the edited genome is probed with primer pairs that amplify donor–vector backbone junction fragments, such as M13F/R1 and M13R/ R2. Genomes that likely harbor only precisely edited alleles are finally analyzed by amplification of the entire edited region, using the F1 and R2 primers, which are complementary to host genomic sequences and not present in the donor DNA. The size of the F1/R2 amplicon should be consistent with that expected from a simple HR-mediated gene-editing event, and the accuracy of the editing event should be verified by sequencing the entire amplicon.