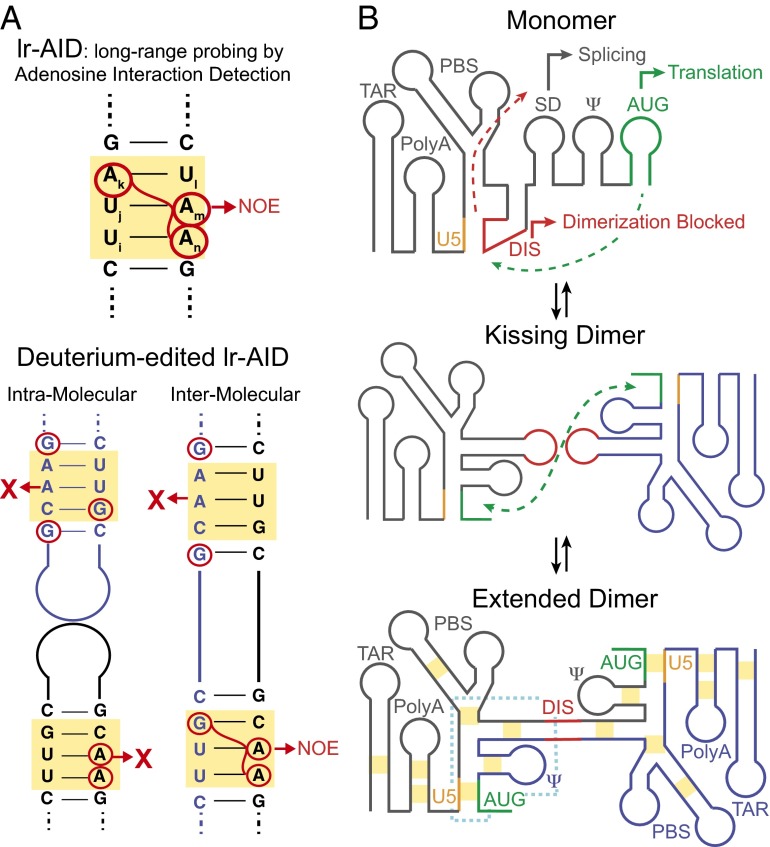
Fig. 1.
New NMR strategy allows characterization of the dimeric interface in the HIV 5′-leader. (A) lr-AID relies on the unique NMR chemical shifts produced by specific base-pair sequences to resolve signals from congested NMR spectra of large RNAs. By differentially silencing all residues other than adenosine or guanosine with deuterium, the deuterium-edited lr-AID approach can distinguish between inter- or intramolecular base-pairing interactions. (B) Proposed mechanism of HIV leader dimerization. In the monomer, the SD and AUG are exposed to promote splicing and translation, respectively. Conversely, the DIS is sequestered by U5, preventing dimerization. In the dimer, DIS dimerization is extended and intermolecular U5:AUG interactions are formed. Yellow boxes indicate the regions probed by deuterium-edited lr-AID and the dotted blue line indicates the core encapsidation signal. The proposed intermediate features a kissing DIS dimer, which was not observed in the present study possibly because it is too low populated and short-lived.