Fig. 6.
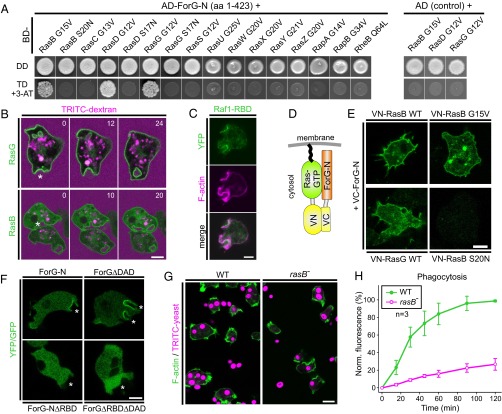
ForG interacts with active Ras. (A, Left) ForG-N interacts specifically with the active forms of RasB, RasG, and RasD in the Y2H assay. Yeast was transformed with the indicated constructs and selected for the presence of prey and bait plasmids by growth on double-dropout (DD) media lacking leucine and tryptophan. Interactions were assayed by growth on stringent triple-dropout (TD) media additionally lacking histidine in the presence of 3 mM 3-AT. (A, Right) Three identified Ras proteins showed no genetic interaction in experiments using the dominant-negative Ras variants or empty AD plasmids as negative controls. AD, Gal4-activation domain; 3-AT, 3-amino-1,2,4-triazole; BD, Gal4-binding domain. (B) YFP-tagged RasG and RasB also localize to macropinosomes during the uptake of TRITC-dextran–labeled medium. Confocal sections are shown and correspond to Movies S5 and S6. Time is given in seconds. Macropinosomes are indicated by stars. (C) Macropinosomes contain active Ras proteins as evidenced by accumulation of the pan-Ras probe Raf1-RBD. A confocal section of a live-cell coexpressing YFP-tagged Raf1-RBD and the F-actin probe LifeAct-mRFP is shown. (D) Scheme of the BiFC strategy. VC, Venus C-terminal fragment; VN, Venus N-terminal fragment. (E) ForG-N interacted with active or WT forms of RasB and RasG, but not with the dominant-negative control of RasB in the BiFC assay. Representative confocal sections are shown. (F) Removal of the putative RBD in ForG-N and the constitutively active ForGΔDAD variant abolished localization of the fusion proteins at endocytic cups (white stars). (G) Strongly impaired yeast phagocytosis of rasB− cells. Accumulation of TRITC-labeled yeast particles by phagocytosis after 45 min of incubation on glass coverslips. Confocal sections are shown. (H) Quantification of phagocytosis with TRITC-labeled yeast revealed strikingly reduced uptake of large particles (mean ± SD, n = 3 for each cell line). Norm., normalized. (Scale bars: B–F, 5 μm; G, 10 μm.)