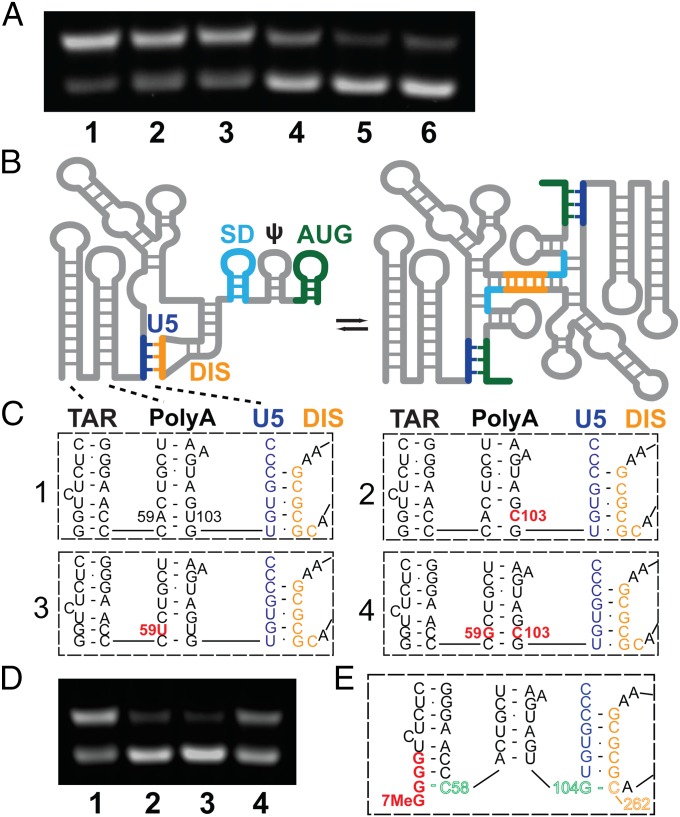
Fig. 4.
(A) Influence of 5′-guanosine number and capping on dimerization, as follows: lane 1: 1G 5′-L RNA; lane 2: Cap1G 5′-L; lane 3: 2G 5′-L; lane4: Cap2G 5′-L; lane 5: 3G 5′-L; and lane 6: Cap3G 5′-L. Under the conditions used, RNAs in lanes 1–3 favor the dimer, whereas those in lanes 4–6 favor the monomer. (B–D) Mutations engineered to destabilize the base of PolyA favor the monomer. (B) Working model for secondary structural changes associated with the monomer–dimer equilibrium (adapted from ref. 26). (C) Proposed base pairing of residues in the lower stems of the TAR and PolyA hairpins and the U5:DIS pseudoknot for wild-type (1), U103C (2), A59U (3), and A59G/U103C (4) constructs. (D) Influence of point mutations on dimerization; lane numbers correspond to the panel labels in C. (E) Model for TSS-dependent dimerization control. In Cap3G transcripts, G(−1)–C58 base pairing disrupts and remodels the PolyA hairpin, allowing G104 to base pair with C262 of the DIS palindrome and thereby stabilizing the monomeric conformer.