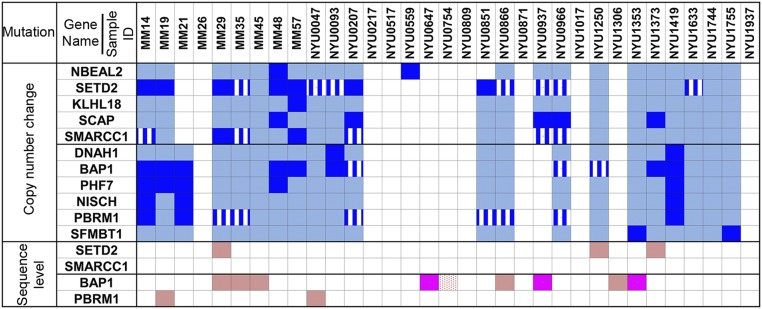
Fig. 2.
Summary of genomic alterations in 33 malignant mesothelioma cases. Each column indicates genomic alterations in each case. Genes NBEAL2, SETD2, KLHL18, SCAP, and SMARCC1 comprise gene cluster 1; genes DNAH1, BAP1, PHF7, NISCH, PBRM1, and SFMBT1 comprise gene cluster 2. Biallelic deletion detected by both, high-density aCGH array and tNGS, are shown in dark blue. Monoallelic loss detected by both, high-density aCGH array and tNGS, are shown in light blue. Blue with white lines represents biallelic deletion detected by tNGS only. Bright pink: two separate somatic mutations of the same gene detected by tNGS at sequence level; light pink: one somatic mutation detected by tNGS at sequence level; dotted pink: one germ-line and one somatic mutation detected by tNGS at sequence level.