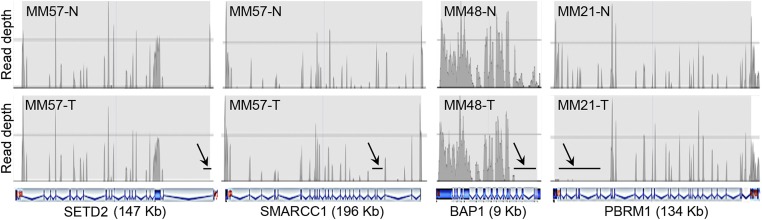
Fig. 3.
CN loss detected in SETD2, SMARCC1, BAP1, and PBRM1 by tNGS. The read depth of each target region was measured and compared by paired analysis between tumor DNA and its normal DNA extracted from blood of the same case. (Upper) The read patterns of the normal DNA for the indicated genes; (Lower) read patterns of the tumor DNAs. Arrows and black horizontal bars indicate areas showing no read corresponding to deleted regions of the gene. Schematics of introexon patterns, characteristic of each gene, shown in blue, are beneath the sample profiles. MM57 shows deletions in exon 1 of SETD2 and exons 4–5 of SMARCC1; MM48 shows deletions in exons 1–5 of BAP1 genes; MM21 shows deletions in exons 22–30 of PBRM1 gene. Estimated CNs are reported in Table S4.