Significance
Most organisms developed a circadian clock to adapt their behavior to daily changes of light and temperature. The molecular clock is remarkably conserved across species with much of our current understanding coming from Drosophila studies. To generate circadian behavior, appropriate levels of neuronal electrical activity are crucial, but the regulators of this activity have remained largely elusive. Here we identify three membrane proteins that interact to set the clock neurons to “day” or “night,” forming a light-input pathway to the circadian clock. The membrane-anchored extracellular protein Quasimodo affects both the daily changes in physiological properties and light responses of brain clock neurons, possibly acting upstream of the potassium channel Shaw and the Na+, K+, Cl− ion transporter NKCC.
Keywords: circadian rhythms, light input, membrane excitability, GABA reversal potential, potassium currents
Abstract
We have characterized a light-input pathway regulating Drosophila clock neuron excitability. The molecular clock drives rhythmic electrical excitability of clock neurons, and we show that the recently discovered light-input factor Quasimodo (Qsm) regulates this variation, presumably via an Na+, K+, Cl− cotransporter (NKCC) and the Shaw K+ channel (dKV3.1). Because of light-dependent degradation of the clock protein Timeless (Tim), constant illumination (LL) leads to a breakdown of molecular and behavioral rhythms. Both overexpression (OX) and knockdown (RNAi) of qsm, NKCC, or Shaw led to robust LL rhythmicity. Whole-cell recordings of the large ventral lateral neurons (l-LNv) showed that altering Qsm levels reduced the daily variation in neuronal activity: qsmOX led to a constitutive less active, night-like state, and qsmRNAi led to a more active, day-like state. Qsm also affected daily changes in K+ currents and the GABA reversal potential, suggesting a role in modifying membrane currents and GABA responses in a daily fashion, potentially modulating light arousal and input to the clock. When directly challenged with blue light, wild-type l-LNvs responded with increased firing at night and no net response during the day, whereas altering Qsm, NKKC, or Shaw levels abolished these day/night differences. Finally, coexpression of ShawOX and NKCCRNAi in a qsm mutant background restored LL-induced behavioral arrhythmicity and wild-type neuronal activity patterns, suggesting that the three genes operate in the same pathway. We propose that Qsm affects both daily and acute light effects in l-LNvs probably acting on Shaw and NKCC.
All organisms are subject to predictable but drastic daily environmental changes caused by the earth’s rotation around the sun. It is critical for the fitness and well-being of an individual to anticipate these changes, and this anticipation is done by circadian timekeeping systems (clocks). These regulate changes in behavior, physiology, and metabolism to ensure they occur at certain times during the day, thereby adapting the organism to its environment (1). The circadian system consists of three elements: the circadian clock to keep time, inputs that allow entrainment, and outputs that influence physiology and behavior (2). Like a normal clock, circadian clocks run at a steady pace (24 h) and can be reset. In nature this environmental synchronization is done via daily light and temperature cycles, food intake, and social interactions (3).
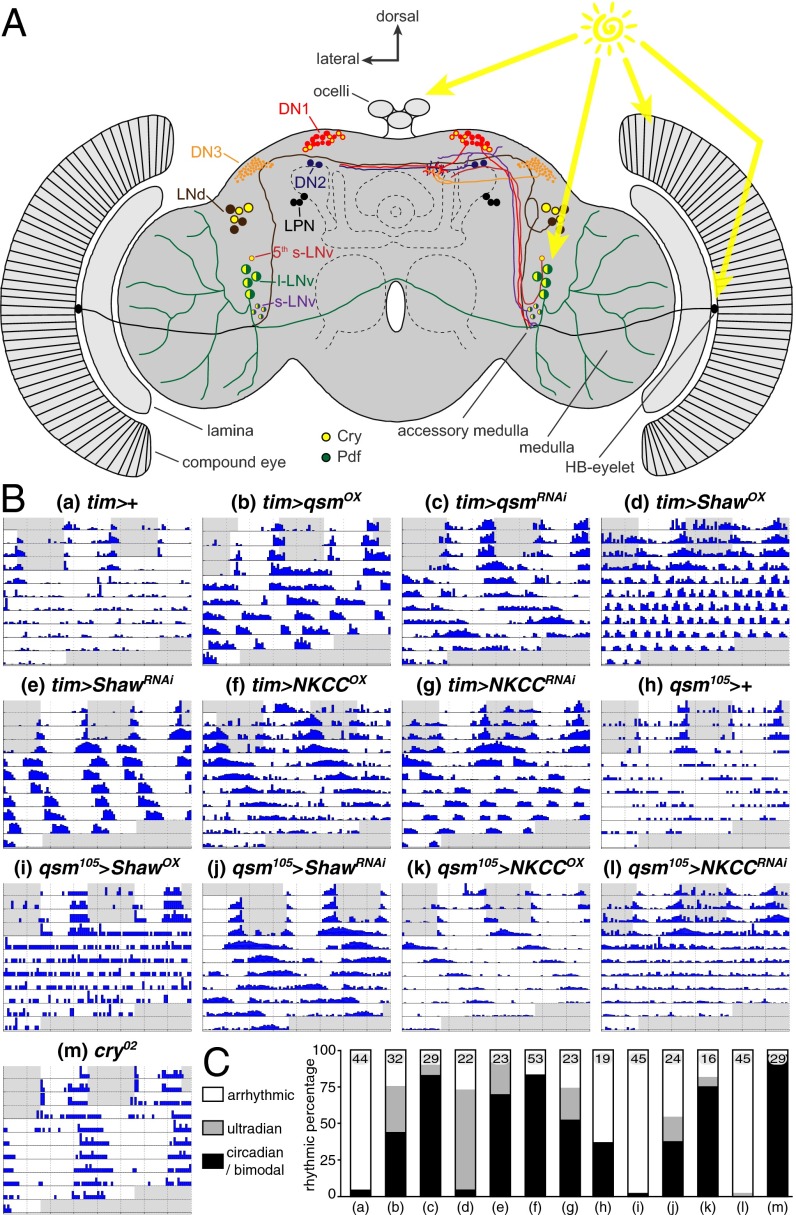
In Drosophila the central clock comprises 75 neuron pairs grouped into identifiable clusters that subserve different circadian functions (Fig. 1A). The molecular basis of the circadian clock is remarkably conserved from Drosophila to mammals (4). This intracellular molecular clock drives clock neurons to express circadian rhythms in electrical excitability, including variation in membrane potential and spike firing. Clock neurons are depolarized and fire more during the day than at night, and circadian changes in the expression of clock-controlled genes encoding membrane proteins such as ion channels and transporters likely contribute to these rhythms (5–8). Such cyclical variations in activity play a critical role in synchronizing different clock neurons and conveying circadian signals to other parts of the nervous system and body (9, 10). Furthermore, they provide positive feedback to the molecular clock, which can dampen rapidly without such feedback (7, 11, 12).
Fig. 1.
The Drosophila circadian clock and LL behavior. (A) Cartoon of the Drosophila brain showing the ∼75 pairs of clock neurons forming several groups [six dorsal lateral neurons (LNd); four large and four small Pdf-expressing ventral lateral neurons (LNv) and a single Pdf-negative neuron (fifth s-LNv); three lateral posterior neurons (LPN); and dorsal neurons (16 DN1, 2 DN2, 40 DN3)] and the potential light input pathways (yellow arrows) via classical photoreceptors, ocelli, the HB-eyelet, and Cry+ neurons. (B) Double-plotted actograms of exemplary individual flies of the indicated genotypes recorded for the first 3 d in a 12-h:12-h light/dark cycle (LD) (gray, lights off; white, lights on) and then in LL. (C) Bar graph specifying the percentage of flies showing circadian or bimodal (black bars), ultradian (light gray bars), or arrhythmic (open bars) behavior for each genotype as indicated by small letters referencing the subpanels in B. Numbers in bars state the number of flies of each genotype.
Light resets the circadian clock every morning to synchronize the clock to the environment via Timeless (Tim) degradation after activation of the blue-light photoreceptor Cryptochrome (Cry), Quasimodo (Qsm), and potentially also visual photoreceptors (13–17). Qsm acts either independently or downstream of Cry and also is able to affect clock protein stability in Qsm-negative neurons by an unknown non–cell-autonomous mechanism (16). Recently Cry has been shown to regulate clock neuron excitability via the redox sensor of the Hyperkinetic voltage-gated potassium (KV)-β subunit (Hk) (18, 19), and here we ask if Qsm affects the clock neurons in a similar way.
Membrane potential is important for control of circadian behavior, and manipulation of Shaw and the Narrow Abdomen (NA) channels, both of which are expressed and function within clock neurons influence neuronal electrical activity, the circadian clock, and clock-controlled behavior in both flies (20–22) and mice (23–25). The firing rate is a key component in mammalian circadian rhythmicity and can be regulated by regional and circadian expression of the sodium potassium chloride cotransporter NKCC, which switches the effects of GABA from inhibitory to excitatory across the day (26, 27).
Here we show that down-regulation or overexpression of the three membrane proteins encoded by the genes qsm, Shaw, and NKCC leads to rhythmicity in constant illumination (LL) and that these genes interact. All three genes are expressed in the well-characterized pigment-dispersing factor (Pdf)- and Cry-positive large ventral lateral neurons (l-LNv) (21, 28), which are important for arousal and light input to the clock (18, 19, 29–32). We used whole-cell recordings of l-LNvs to characterize their physiological properties and acute light effects across the day and found that Qsm helps set the circadian state of clock neurons and modifies their response to light, possibly by acting via Shaw and NKCC.
Results
Rhythmic LL Behavior and the Interaction of Qsm with Shaw and NKCC.
Light resets the circadian clock, and consequently, wild-type flies displayed arrhythmic behavior in constant dim white light (LL, 10 µW/cm2), whereas cry02 loss-of-function mutants were rhythmic (Fig. 1 B and C and Table S1) (15, 33). Previously we have shown that Qsm triggers Tim degradation within clock cells, and knockdown of qsm [qsmRNAi, i.e., >70% reduction of qsm mRNA (16)] in all clock neurons using tim-gal4 resulted in robust long-period (∼27-h) locomotor rhythms in LL (16). We confirmed that qsmRNAi induces robust and long-period rhythms in LL using two independent qsm-RNAi lines. Interestingly, qsm overexpression (qsmOX) led to shorter, ∼13-h rhythms, most likely reflecting the persistence of morning and evening activity peaks in LL.
Table S1.
Circadian behavior of flies in LL
| Genotype | tim allele | % rhythmic | Period, h (SD) | Rhythmic statistics | n |
| tim > + (wild-type) | ls | 4.5 | — | — | 44 |
| tim > qsmOX | ls | 75.0 | 13.4 (1.8) | 2.2 (0.7) | 32 |
| tim > qsmRNAi | ls | 89.7 | 26.6 (1.6) | 2.4 (0.6) | 29 |
| tim > qsmRNAi(2) | ls | 98.1 | 27.6 (2.0) | 3.3 (1.0) | 104 |
| tim > ShawOX | ls | 72.7 | 4.2 (0.6) | * | 22 |
| tim > ShawRNAi | ls | 91.3 | 12.5 (0.8) | 2.0 (0.7) | 23 |
| tim > NKCCOX | ls | 83.0 | 28.9 (0.8) | 2.4 (1.0) | 53 |
| tim > NKCCRNAi | ls | 73.9 | 13.3 (1.1) | 2.3 (0.8) | 23 |
| tim > qsmOX | s | 34.0 | 30.3 (5.6) | 2.1 (0.4) | 50 |
| tim > qsmRNAi | s | 31.4 | 30.2 (5.4) | 2.5 (1.2) | 35 |
| tim > qsmRNAi(2) | s | 40.0 | 24.6 (2.5) | 2.3 (0.6) | 20 |
| tim > ShawRNAi | s | 58.3 | 29.8 (5.4) | 2.5 (1.2) | 24 |
| tim > NKCCOX | s | 26.6 | 34.1 (3.9) | 2.4 (1.2) | 30 |
| cry02 | s | 96.6 | 24.3 (1.3) | 3.1 (1.0) | 29 |
| gl60j | ls | 0 | – | – | 22 |
| norpAP41 | s | 0 | – | – | 32 |
| HdcJK910 | s/ls | 16.6 | 25.6 (4.3) | 2.3 (0.8) | 36 |
| qsm105 > + | ls | 36.8 | 27.4 (1.9) | 2.4 (0.8) | 19 |
| qsm105 > ShawOX | ls | 2.2 | – | – | 45 |
| qsm105 > ShawRNAi | ls | 54.2 | 26.6 (2.9) | 2.3 (0.7) | 24 |
| qsm105 > NKCCOX | ls | 81.3 | 29.1 (1.2) | 3.3 (0.8) | 16 |
| qsm105 > NKCCRNAi | ls | 2.2 | – | – | 45 |
This table is related to Fig. 1 and Fig. S2. For each genotype, the percentage of the overall rhythmic flies, the mean (SD) of the prevalent rhythmic category, the rhythmic statistics, and the number of flies, n, are given.
Rhythmic statistics values could not be calculated for periods of this length using the signal-processing tool-box in MatLab. Instead, rhythmicity for each fly was deemed significant based on periodogram analysis using Actogram J with a P value cut-off set to 0.001. Each period was subsequently verified by manual inspection of individual actograms.
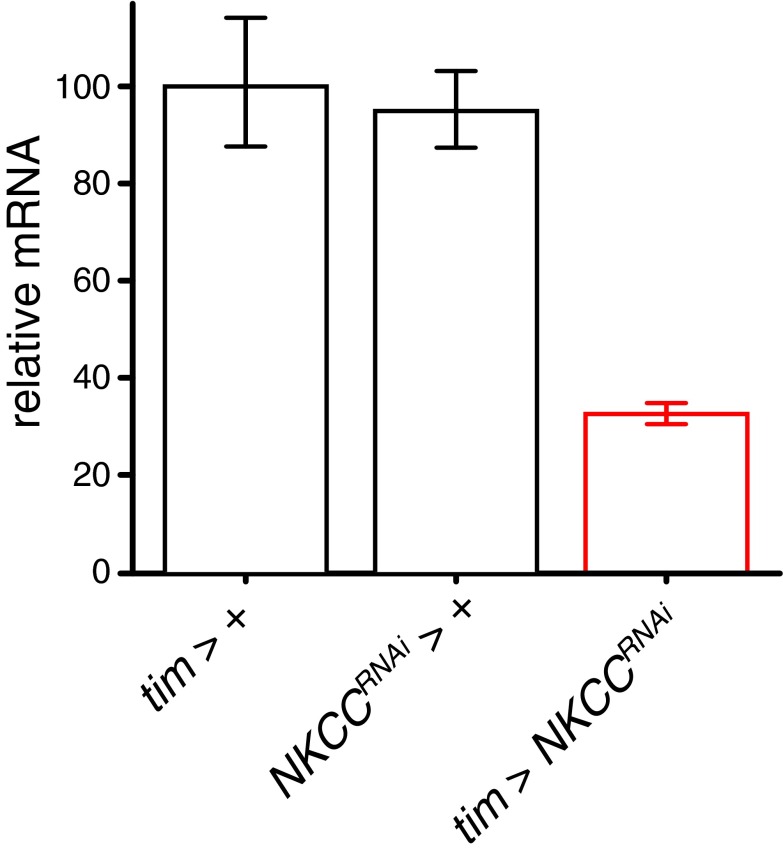
Because NKCC interacts physically with Qsm in yeast (34) and because Shaw expression in clock neurons partially overlaps with the expression of qsm (16, 21), we tested whether NKCC and Shaw would also affect LL behavior. Changing levels of both NKCC and Shaw resulted in flies that were rhythmic in dim LL. Like qsmRNAi flies, flies overexpressing NKCC (NKCCOX flies) exhibited long periods (∼29 h), whereas NKCC knockdown [NKCCRNAi ; an ∼70% reduction of mRNA (Fig. S1)] and Shaw knockdown [ShawRNAi ; an ∼90% reduction of Shaw (29)] resulted in ∼13-h periods, reflecting potential bimodal behavior similar to that of qsmOX. Shaw overexpression (ShawOX) induced remarkably robust ultradian rhythms with an ∼4-h period, indicating compromised circadian clock function (21) and uncovering an underlying ultradian rhythm presumably generated by a membrane-based oscillator. These manipulations did not affect behavior in constant darkness (DD) (Table S2), with the exception of ShawOX, which led to arrhythmicity as previously reported (21).
Fig. S1.
RT-qPCR verification of NKCCRNAi knockdown (related to Figs. 1–5). qPCR analysis of relative mRNA levels in whole heads showing the efficiency of RNAi-mediated knockdown of NKCC (tim > NKCCRNAi) compared with controls (either gal4 or uas alone). Bars represent means; whiskers represent minimum and maximum.
Table S2.
Circadian behavior of flies in DD
| Genotype | tim allele | % rhythmic | Period, h (SD) | Rhythmic statistics | n |
| tim > + (wild-type) | ls | 93.8 | 23.6 (0.4) | 2.8 (0.8) | 16 |
| tim > qsmOX | ls | 100 | 24.1 (0.2) | 3.9 (0.4) | 8 |
| tim > qsmRNAi | ls | 100 | 24.1 (0.2) | 4.0 (0.4) | 8 |
| tim > ShawOX* | ls | 13 | 24.4 (0.4) | 2.5 (0.2) | 39 |
| tim > ShawRNAi* | ls | 97 | 24.3 (0.1) | 4.8 (0.3) | 32 |
| tim > NKCCOX | ls | 85.7 | 24.2 (0.3) | 2.9 (0.6) | 14 |
| tim > NKCCRNAi | ls | 81.3 | 24.6 (0.5) | 2.7 (0.7) | 16 |
The light sensitivity of wild-type flies is also influenced by the naturally occurring s-tim/ls-tim polymorphism (35, 36), and we therefore performed experiments with both alleles. In all tested cases (qsmOX, qsmRNAi, ShawRNAi, and NKCCOX), s-tim caused a dramatic (∼50%) reduction in the percentage of rhythmic flies compared with ls-tim flies and often was correlated with period lengthening (Fig. S2 and Table S1). This result fits well with the previous observation that flies carrying the s-tim allele are more light sensitive than their ls-tim counterparts, and the dramatic behavioral difference caused by the two alleles underscores their biological importance (35, 36).
Fig. S2.
LL behavior of flies carrying the s-tim allele (related to Fig.1 and Table S1). Double-plotted actograms of exemplary individual flies of the indicated genotypes recorded for the first 4 d in LD (gray, lights off; white, lights on) followed by 9 d of constant dim light.
Because the tim-gal4 driver is also expressed in the photoreceptor cells of the compound eyes, we also tested flies with impaired visual system function. Similar to wild-type flies, flies lacking (i) all external photoreceptors (gl60j) (15), or (ii) phospholipase C-β required for the visual phototransduction cascade (norpAP41) (37), or (iii) histamine, the principal neurotransmitter of the visual system (HdcJK910) (38), became virtually arrhythmic during LL (Table S1). These results strongly suggest that the tim-gal4–mediated LL rhythmicity is induced by the alteration of qsm, Shaw, and NKCC expression within the clock neuronal network.
To test directly for genetic interactions, we determined the effects of altering Shaw and NKCC levels on the LL-rhythmic phenotype induced by reducing Qsm. We used qsm105, an intragenic gal4 insertion line that reduces qsm expression, and induced ∼37% LL rhythmicity in heterozygous (qsm105 > +) flies (Fig. 1 B and C and Table S1) (16). With the qsm105 driver, both ShawOX and NKCCRNAi induced wild-type scores of LL rhythmicity (i.e., 2%), whereas the opposite manipulation led to increased rhythmicity scores (54% and 81%, respectively). The apparent genetic interaction among qsm, Shaw, and NKCC suggests that they function in the same pathway. We propose that Qsm influences membrane properties via ion channels (Shaw) and transporter proteins (NKCC) that in turn might influence the molecular responses of circadian clock neurons to light.
Qsm Affects Daily Changes in Physiological Properties.
To determine how Qsm, Shaw, and NKCC might contribute to clock neuronal activity, we examined their neurophysiological roles by performing whole-cell recordings from l-LNv with altered qsm, Shaw, or NKCC expression using Pdf-gal4 and uas-RFP (Fig. S3 A and B). We focused on the l-LNv because they represent the electrophysiologically best-characterized clock neuronal cell type and because they express qsm, Shaw, and NKCC (8, 21, 28).
Fig. S3.
Whole-cell recording preparation and response of l-LNvs to acute light (related to Figs. 2, 4, and 5 and Table S2). (A) Whole-brain preparation showing the Pdf-RFP–labeled l-LNvs for recording (the band in the middle is a nylon thread holding the brain in the recording chamber). (B) Detail of the l-LNvs with the recording electrode (viewed from below). (C) In wild-type control flies green light (555 nm) had no effect on wild-type l-LNvs either in the daytime or at nighttime. (D) cry02 l-LNvs did not respond to blue light (470 nm) with a change of firing either in the daytime or at nighttime. (E–H) l-LNvs with altered Shaw or NKCC levels (Pdf-gal4) showed no differential response in recordings taken during the daytime (Left) or at nighttime (Right). Note that the scale of the graphs in G differs from the scale used in the graphs in the other panels. In each panel the traces show an example of a current-clamp recording of an l-LNv for 1 min before, 30 s during, and 2.5 min after exposure to light (indicated by the green or blue bar). The graphs show a quantification of the light response from multiple recordings. N.B.: In the genotypes not responding to acute light (qsmOX, ShawOX, NKCCRNAi), 18 neurons did not spike or spiked only very occasionally; the neurons that did spike regularly did not change their firing rate. However, some neurons (n = 8) showed a startle response with briefly increased or decreased spiking (e.g., left panels in F and Fig. 4C). The examples shown are from occasionally spiking neurons. In the graphs the solid line shows the mean, and the gray shading shows the SD. The number of recorded neurons of each genotype is indicated.
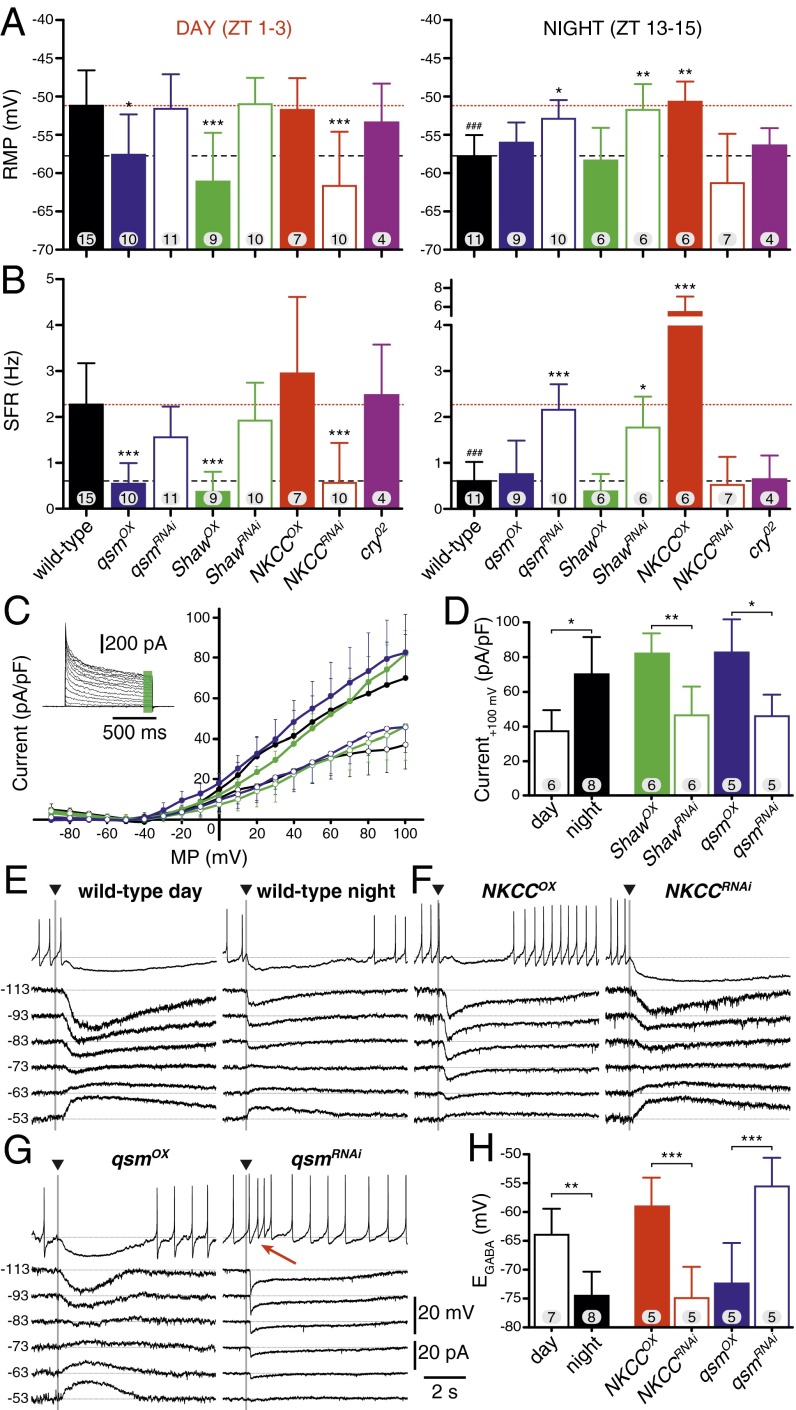
Although physiological parameters showed variability among recordings of different animals, wild-type l-LNvs showed a more depolarized resting membrane potential (RMP) and a higher spontaneous firing rate (SFR) in the day than at night (Fig. 2 A and B and Table S3) (5, 31). In daytime recordings, qsmOX led to a more hyperpolarized RMP and reduced SFR that were very similar to control and qsmOX nighttime recordings. In contrast, qsmRNAi depolarized the neurons and increased SFR at night to levels similar to those in wild-type and qsmRNAi l-LNvs. We tested two qsmRNAi lines targeting different regions of the qsm transcript (16) and did not observe a difference between the two lines (pooled data are shown). Interestingly, the differences in daytime and nighttime RMP and SFR in wild-type l-LNvs were abolished in both qsmOX and qsmRNAi l-LNvs. Resembling the changes seen for qsm, ShawOX led to more hyperpolarized, less active neurons, and ShawRNAi led to more depolarized and more actively firing neurons. NKCC manipulations had the opposite effect, with NKCCOX depolarizing and activating the neurons, in particular at nighttime, and with NKCCRNAi hyperpolarizing and inactivating the neurons.
Fig. 2.
Daily changes in the physiological properties of l-LNvs. (A) Changes in the RMP of wild-type neurons (black bars) and l-LNvs with altered levels (Pdf-gal4) of qsm (blue bars), Shaw (green bars), and NKCC (red bars), and cry02 mutants (purple bars) in the day (Left) and at night (Right). Solid colored bars represent overexpression and open bars indicate RNAi knockdown of the respective genes. Data are presented as means; whiskers indicate the SD; the numbers in bars state the numbers of neurons for each genotype. The dotted red line represents wild-type daytime values, and the dashed black line indicates wild-type nighttime values. (B) SFR of the genotypes in A. (C) Current densities (measured at time point indicated by the green bar in the example shown in the inset) for wild-type l-LNvs in the daytime (open black circles) and nighttime (closed black circles), ShawOX l-LNvs (closed green circles), ShawRNAi l-LNvs (open green circles), qsmOX l-LNvs (closed blue circles), and qsmRNAi l-LNvs (open blue circles). Data are presented as means; whiskers indicate the SD. (D) Quantification of the current density at +100 mV. (E–G) Representative examples of the GABA response of a wild-type l-LNv (E), NKCCOX and NKCCRNAi (F), and qsmOX and qsmRNAi (G) during the daytime (Left) and nighttime (Right). The top trace in each panel is the current-clamp response to GABA (10 ms, 25 mM, 10 psi) at the arrowhead. Bottom traces show the same neuron and the GABA pulse in voltage clamp held at the indicated potentials. Note the red arrow in G pointing to occasional excitatory GABA effects for qsmRNAi. (H) Quantification of EGABA for the indicated genotypes. *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA with Dunnett’s (A and B) test or Tukey’s (D and H) post hoc test (*) or t test (#).
Table S3.
Physiological parameters (RMP, SFR) and acute light response (Fon − Foff) in daytime and nighttime
| Genotype | Day or night | RMP, mV (SD) | SFR, Hz (SD) | Fon − Foff, Hz (SD) | n |
| Pdf > + (wild-type) | Day | −51.2 (4.6) | 2.27 (0.90) | −0.23 (0.84) | 15 |
| Night | −57.7 (2.7) | 0.60 (0.41) | 1.61 (1.17) | 11 | |
| Pdf > qsmOX | Day | −57.6 (5.3) | 0.54 (0.46) | 0.17 (0.22) | 10 |
| Night | −56.0 (2.6) | 0.74 (0.73) | 0.06 (0.52) | 9 | |
| Pdf > qsmRNAi | Day | −51.6 (4.5) | 1.56 (0.67) | 1.50 (1.31) | 11 |
| Night | −52.9 (2.5) | 2.15 (0.56) | 1.41 (0.98) | 10 | |
| Pdf > ShawOX | Day | −61.1 (6.3) | 0.34 (0.47) | −0.02 (0.17) | 9 |
| Night | −58.3 (4.3) | 0.35 (0.39) | −0.06 (0.14) | 6 | |
| Pdf > ShawRNAi | Day | −51.0 (3.4) | 1.93 (0.82) | 1.56 (1.61) | 10 |
| Night | −51.7 (3.3) | 1.76 (0.68) | 1.21 (0.69) | 6 | |
| Pdf > NKCCOX | Day | −51.7 (4.1) | 2.94 (1.66) | 1.52 (0.99) | 7 |
| Night | −50.7 (2.7) | 5.30 (1.74) | 1.20 (1.14) | 6 | |
| Pdf > NKCCRNAi | Day | −61.7 (7.1) | 0.56 (0.87) | 0.11 (0.15) | 10 |
| Night | −61.3 (6.4) | 0.51 (0.61) | −0.02 (0.21) | 7 | |
| cry02 | Day | −53.3 (5.1) | 2.47 (1.11) | −0.06 (0.38) | 4 |
| Night | −56.5 (2.4) | 0.63 (0.53) | 0.11 (0.19) | 4 | |
| tim > qsmRNAi/GFP | Day | −54.9 (2.7) | 5.78 (0.84) | – | 9 |
| Night | −55.8 (2.1) | 5.70 (1.26) | – | 8 | |
| tim > qsmRNAi/ShawOX | Day | −61.2 (3.1) | 0.47 (0.57) | – | 9 |
| Night | −59.0 (2.5) | 0.90 (0.84) | – | 7 | |
| tim > qsmRNAi/NKCCRNAi | Day | −57.6 (1.8) | 1.90 (0.72) | – | 7 |
| Night | −55.7 (3.5) | 2.56 (0.56) | – | 7 |
Qsm Affects Daily Changes in K+ Currents and the GABA Reversal Potential, and Qsm Interacts with Shaw and NKCC.
To elucidate potential mechanisms by which Qsm could affect physiological properties, we tested whether the effect might be caused by alterations in voltage-dependent somatic currents affecting the RMP. We examined the current–voltage relationships of l-LNvs and measured the non-inactivating currents that include Shaw (Fig. 2 C and D and Table S4) (39, 40). We analyzed the sustained current densities of wild-type l-LNvs at daytime and nighttime and could not detect any difference around the RMP, most likely because of masking by the leak-current subtraction. There was a difference, however, at positive holding potentials (+100 mV), with current densities at night being >30 pA/pF larger than in the day, suggesting more potassium channels were open at night. ShawOX resulted in a high current density similar to that recorded in wild-type l-LNvs at night, whereas ShawRNAi values matched wild-type daytime recordings, indicating that Shaw is involved in the observed day/night difference. Mimicking Shaw, qsmOX had high current densities similar to those in wild-type l-LNvs at night, and qsmRNAi currents matched wild-type day levels. This observation suggests that Qsm interacts with voltage-gated potassium channels such as Shaw in setting membrane properties.
Table S4.
Sustained current density at +100 mV
| Genotype | Current, pA/pF (SD) | n |
| Pdf > + (wild-type) day | 37.1 (12.0) | 6 |
| Pdf > + (wild-type) night | 69.9 (21.6) | 8 |
| Pdf > ShawOX | 81.9 (11.7) | 6 |
| Pdf > ShawRNAi | 46.1 (16.7) | 6 |
| Pdf > qsmOX | 82.5 (19.2) | 5 |
| Pdf > qsmRNAi | 45.8 (12.4) | 5 |
This table is related to Fig. 2. Data are shown as mean (SD). n, the number of measured neurons.
Manipulating NKCC levels had an effect on RMP and SFR, and although NKCC activity is electrically neutral, NKCC pumps chloride ions into the cell, increasing the intracellular chloride concentration. We therefore tested whether the GABA reversal potential (EGABA) of l-LNvs also changes across the day by injecting GABA (10 ms, 25 mM, 10 psi) into the ipsilateral medulla. In all cases spiking activity was affected. We measured the induced currents and calculated EGABA from the resulting I–V curve (Fig. 2 E–H and Table S5). During the day the EGABA was more positive (>10 mV), making GABA input less effective. GABA injections in the daytime or nighttime increased the EGABA in NKCCOX brains and had the opposite effect in NKCCRNAi brains in which the EGABA was more negative, thereby mimicking the wild-type day/night change. Changing Qsm levels also had an effect that again was in the opposite direction of NKCC, with qsmOX decreasing and qsmRNAi increasing the EGABA. Strikingly, in some cases the EGABA was more positive than the RMP and GABA consequently acted as an excitatory neurotransmitter that induced firing (Fig. 2G, red arrow in the top trace). These results suggest that Qsm interacts with NKCC and that they participate in the daily change in GABA efficiency.
Table S5.
EGABA
| Genotype | EGABA, mV (SD) | n |
| Pdf > + (wild-type) day | −63.8 (4.3) | 7 |
| Pdf > + (wild-type) night | −74.6 (4.2) | 8 |
| Pdf > NKCCOX | −59.0 (5.0) | 5 |
| Pdf > NKCCRNAi | −74.8 (5.3) | 5 |
| Pdf > qsmOX | −72.5 (7.2) | 5 |
| Pdf > qsmRNAi | −55.6 (4.9) | 5 |
This table is related to Fig. 2. Data are shown as mean (SD), n, the number of measured neurons.
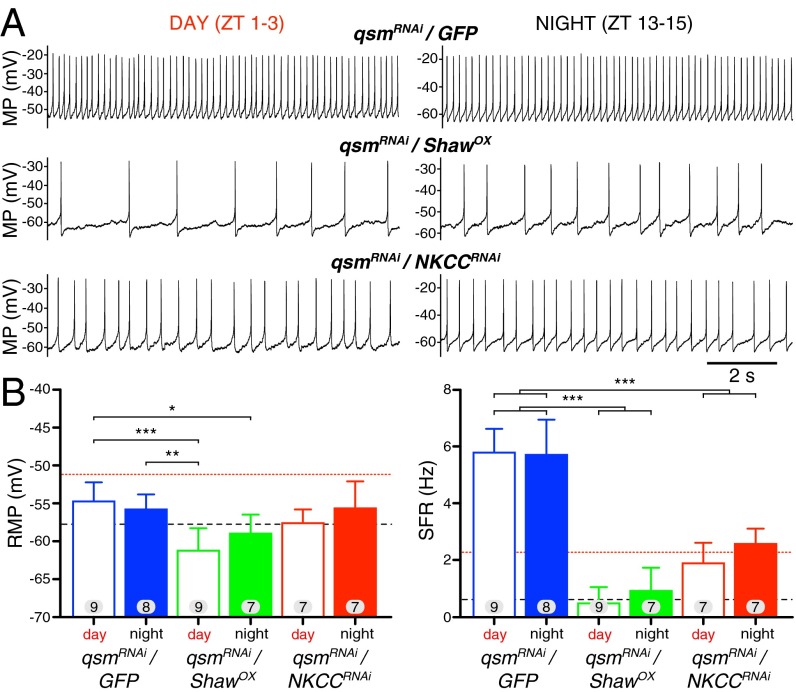
To characterize the interaction of Qsm with Shaw and NKCC further, we tested whether altering levels of Shaw and NKCC could rescue some of the qsm-induced physiological changes. We used tim-gal4 > qsmRNAi to reduce Qsm expression in all clock neurons and found an effect similar to that seen with Pdf-gal4. Again we did not observe a difference between day and night, but the resulting firing rate was much higher (Fig. 3 and Table S3), suggesting additional network effects. We then coexpressed qsmRNAi with ShawOX and NKCCRNAi and could rescue the elevated firing rate to wild-type levels, mimicking the behavioral rescue experiments and further supporting a cell-autonomous interaction of Qsm with Shaw and NKCC. Interestingly, ShawOX seems to be more effective than NKCCRNAi, and the rescue was less pronounced at night.
Fig. 3.
Physiological interaction of qsm with Shaw and NKCC. (A) Representative traces of l-LNvs coexpressing qsmRNAi with GFP (tim-gal4, control) (Top), ShawOX (Middle), and NKCCRNAi (Bottom) in the daytime (Left) and nighttime (Right). MP, membrane potential. (B) Quantification and statistical analysis of RMP (Left) and SFR (Right) showed a rescue of the qsmRNAi phenotype by both ShawOX and NKCCRNAi. For reference, the dotted red line indicates wild-type daytime values, and the dashed black line indicates nighttime values (from Fig. 2 A and B). Data are presented as means; the whiskers indicate SD. The numbers in the bars show the number of recorded neurons for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA with Tukey’s post hoc test.
In summary, overexpression and RNAi knockdown of qsm and Shaw resulted in opposing phenotypes compared with NKCC manipulations. Interestingly, qsmOX or ShawOX and NKCCRNAi promote the less active nighttime state, whereas qsmRNAi, ShawRNAi and NKCCOX push the neurons into the more depolarized daytime state, eliminating the acute differences between daytime and nighttime in all cases. Furthermore, Qsm interacts with Shaw and NKCC to affect potassium currents and the EGABA.
Effects of Acute Light on l-LNvs and Its Modulation by Qsm.
Altering the levels of Qsm, Shaw, and NKCC led to LL rhythmicity, suggesting an impairment of the light input to the circadian clock. Because the visual system mutants became arrhythmic, the alteration of Qsm, Shaw, and NKCC within clock neurons must be responsible for the observed LL rhythmicity. Although l-LNv neurons are unlikely to be mediators of rhythmic behavior in LL (16, 41–43), they are arousal neurons that are activated by blue light via Cry (18). Therefore we tested the response of l-LNvs to light in isolated brains, i.e., in the absence of all canonical photoreceptors including the compound eye, the HB-eyelet, and the ocelli, leaving only deep-brain photosensitive pathways (e.g., Cry) intact.
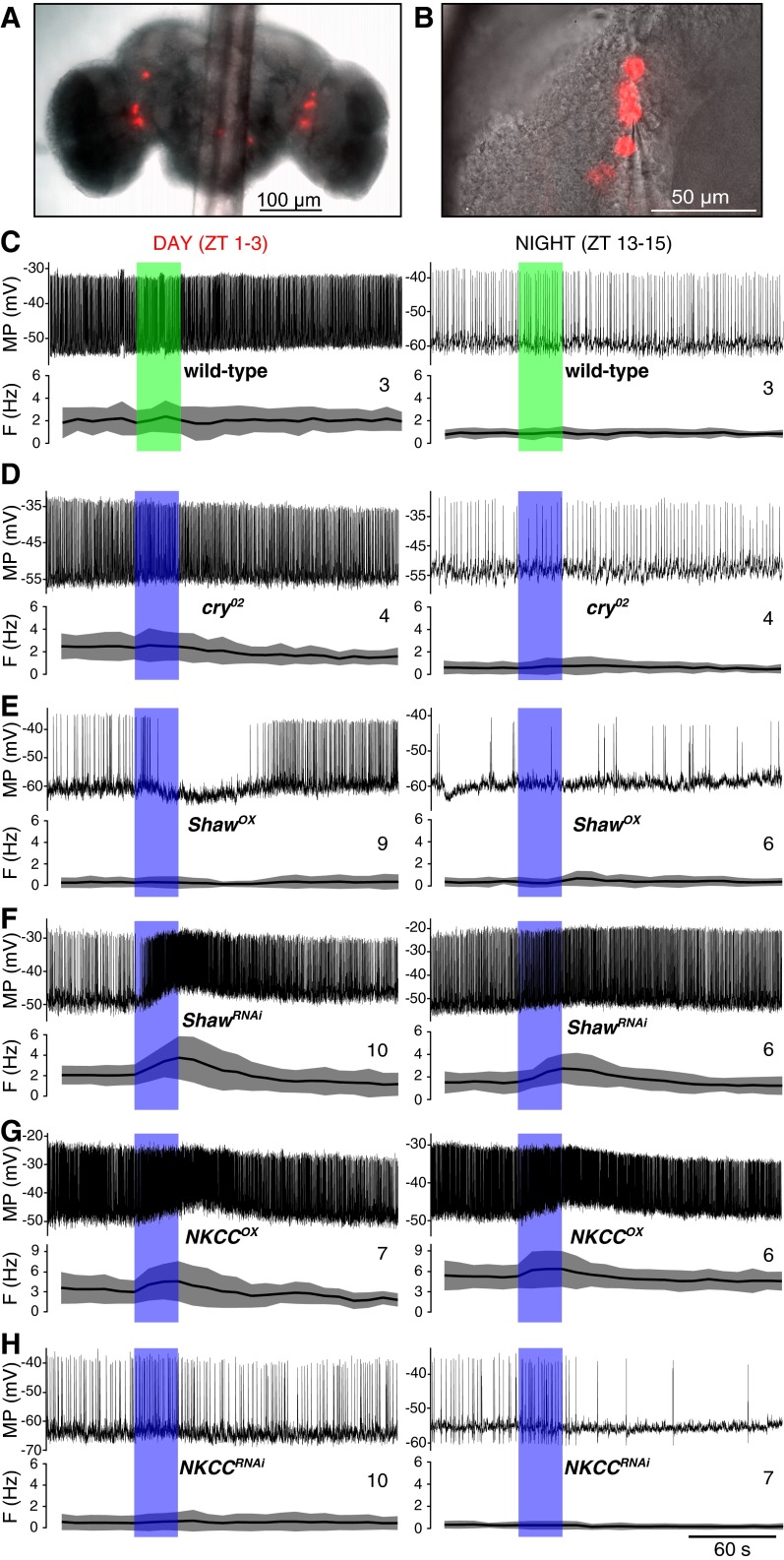
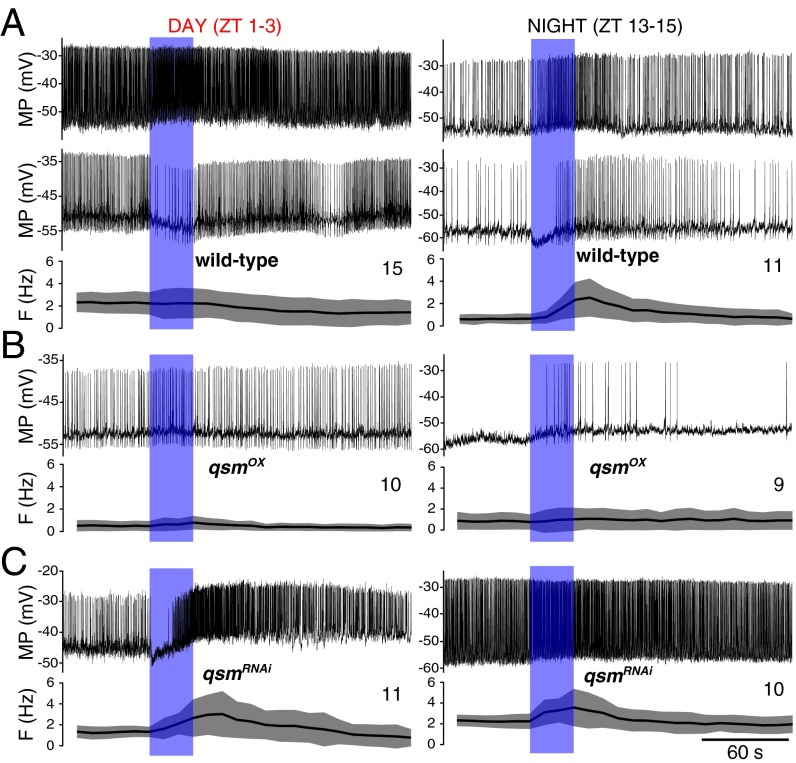
At night all tested wild-type neurons responded to blue light with a slight depolarization and an increase in firing rate, either directly at lights-on (Fig. 4A) or after a brief startle response (Fig. 4A, Bottom), often outlasting the stimulus by tens of seconds before eventually returning to the baseline firing rate. On average, the spiking in response to blue light at night increased by 1–2 Hz to levels near the resting firing rate in the daytime (Fig. 5A and Table S3). In the daytime the response was different, more complex, and variable. Some neurons briefly increased (n = 2) or decreased firing (n = 5), but on average there was not much change in firing frequency, a response clearly different from that observed at night and suggesting daily variation in the responsiveness and light sensitivity of the clock. We occasionally (n = 8) observed a startle response with a brief period of either increased firing or no firing immediately after the lights-on transition both at day and night. Consistent with published data (18), the effect was seen only with blue (470 nm) and not with green (555 nm) (Fig. S3C) or red (625 nm) light. cry02 l-LNvs did not respond to blue light (Fig. S3D) but had day/night changes in RMP and SFR similar to those in control l-LNv, indicating that Cry is not required for the regulation of neuronal electrical activity by the circadian clock.
Fig. 4.
Acute blue-light response of l-LNvs in daytime and nighttime. (A) Response of wild-type neurons to blue light in the daytime (Left) and nighttime (Right). ZT, zeitgeber time. (B and C) l-LNvs with altered qsm levels (Pdf-gal4) showed no differential response depending on the time of day but did show a differential response depending on the level of expression. (Upper) Examples of traces of a current-clamp recording of a l-LNv for 1 min before, 30 s during, and for 2.5 min after exposure to light (indicated by blue bar). (Lower) Quantification of the light response from multiple recordings. The solid line indicates the mean, and gray shading indicates the SD. The number of recorded neurons for each genotype is indicated. F, firing frequency; MP, membrane potential.
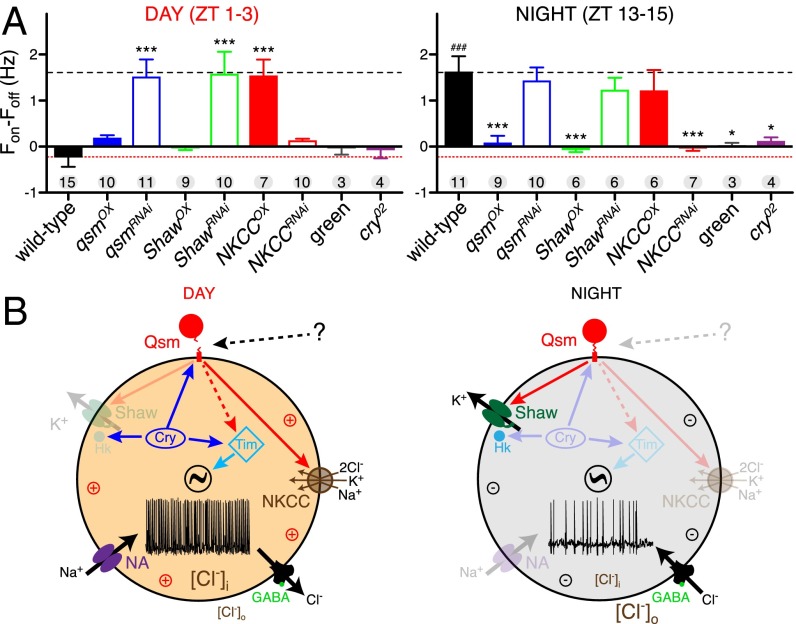
Fig. 5.
The effects of blue light on different genotype l-LNvs and the circadian model. (A) Quantification of the response of the indicated genotypes (Pdf-gal4) to blue light as measured by the activity in the 30-s light on (Fon) period minus the background activity in the 30 s before the light pulse (Foff). Data are shown as means; whiskers indicate SD. The number of recorded neurons for each genotype is indicated. *P < 0.05, ***P < 0.001; one-way ANOVA with Dunnett’s post hoc test (*) or t test (#). (B) Model of l-LNv cellular changes underlying the membrane potential and the firing differences between daytime and nighttime. Qsm receives light input from Cry and affects Shaw and NKCC activity. Additionally, Cry activates Hk and Qsm, and light-activated Cry degrades Tim. Because Qsm is also expressed in Cry-negative clock neurons (16), it is likely that non–cell-autonomous mechanisms also activate this pathway (this possibility is indicated by the question mark). How, and to which extend, Qsm, Shaw and NKCC influence TIM stability is unclear (dashed line). In the daytime, Qsm is cleaved from the membrane, activity of Shaw is low, the sodium leak channel NA is active, and NKCC activity is high, resulting in a more positive RMP and higher spiking rate. In the nighttime Qsm is membrane bound, and Shaw levels are high, but there is less NA and NKCC, resulting in more negative RMP and less spiking.
qsmOX always led to neurons with diminished firing frequency in response to the light pulse (Fig. 4B). Therefore, qsmOX l-LNvs behaved rather like cry02 neurons, the control neurons exposed to green light, or the daytime wild-type neurons. qsmRNAi, in contrast, always led to an increase in the spike rate in response to blue light similar to that seen in wild-type night recordings but starting from a higher baseline rate (Fig. 4C). The average change in firing was very similar to the wild-type nighttime response and was independent of the time of day (Fig. 5A and Table S3). Light pulses in the presence of up- or down-regulation of Shaw resulted in changes similar to those seen with the equivalent alterations of qsm expression, and NKCC manipulations resulted in opposite changes (Fig. S3 E–H). In summary, wild-type flies show a differential day/night response to blue-light pulses, but no such difference was seen in flies with Qsm, Shaw, and NKCC manipulations, the response of these flies being determined instead by the respective up- or down-regulation of gene expression.
Discussion
Light is the dominant circadian zeitgeber that resets the molecular clock. In this study we determined how light affects membrane excitability via the membrane proteins Qsm, Shaw, and NKCC. Previously we have shown that Qsm contributes to circadian clock light input with down-regulation in all clock neurons (tim-gal4) resulting in robust rhythmic behavior in LL (16). Here we show that overexpression (qsmOX) also results in robust LL rhythmicity, but with predominantly ∼13-h periods, suggesting a more robust morning oscillator that is normally weakened in DD conditions (Fig. 1 and Table S1). Manipulating the expression levels of both the potassium channel Shaw and the ion cotransporter NKCC also resulted in LL rhythmicity, whereas several visual system mutants behaved like wild-type flies and became arrhythmic. These experiments show that the membrane proteins encoded by qsm, Shaw, and NKCC control rhythmic behavior in LL. Furthermore, the rescue of wild-type behavior and neurophysiological properties by reciprocal changes of Qsm and Shaw and by the simultaneous reduction of Qsm and NKCC suggests that Qsm interacts genetically and perhaps directly with Shaw and NKCC.
Clock neurons are more depolarized and fire more during the daytime, and circadian changes in the expression of clock-controlled genes such as ion channels and transporters are likely to play a part (7, 8). Contributing to this rhythm is a sodium leak current mediated by NA that recently has been shown to depolarize Drosophila clock neurons (22). Here we show that Shaw, NKCC, and Qsm also contribute to daily electrical activity rhythms: overexpression and RNAi knock-down of qsm and Shaw compared with NKCC resulted in opposing phenotypes. Interestingly, qsmOX or ShawOX and NKCCRNAi promote the less active nighttime state, whereas qsmRNAi, ShawRNAi, and NKCCOX push the neurons into the more depolarized daytime state, eliminating the acute day/night differences in all cases (Fig. 2 and Table S3). We have shown previously that Shaw regulates circadian behavior (21) and that, in agreement with our current findings, Shaw regulates membrane potential and firing in Drosophila motoneurons (29, 44). NKCC activity is electrically neutral but increases the intracellular Cl− concentration so that the GABAA receptor opens in response to GABA but, as a consequence, Cl− presumably exits the cell down its electrochemical gradient, thereby depolarizing the membrane potential so that GABA effectively becomes an excitatory neurotransmitter (6, 12, 26, 29, 45). Our data show that in Drosophila a similar mechanism occurs, which is consistent with potential NKCC enrichment in l-LNv at dawn (28). The mechanism setting the neuronal state to either daytime or nighttime via Qsm, Shaw, and NKCC is likely to be predominantly cell-autonomous, because all components have been shown to act or to be expressed in the l-LNv (8, 21, 28). Although in an earlier study (16) using qsm-gal4 lines we did not detect qsm expression in the l-LNv, these lines may not report expression faithfully in all qsm cells. Now, the finding that qsm RNA is enriched in the l-LNv (28), combined with the strong effects of two qsm-RNAi lines on l-LNv electrical properties presented in this study, indicates that qsm is endogenously expressed in these neurons.
Our physiological studies are limited to the Pdf-expressing l-LNv neurons. These neurons are unlikely candidates for driving behavioral rhythms in LL (41–43), and we showed previously that qsm knockdown in Pdf neurons (s-LNv and l-LNv) does not result in robust LL rhythmicity (16). Therefore the effects of light on the electrical properties of l-LNv reported here do not necessarily explain the LL rhythmicity observed after manipulating qsm, Shaw, and NKCC in all clock neurons. However, the electrophysiological results using tim-gal4 show that Qsm, Shaw, and NKCC could fulfill similar functions in other clock neurons, including those crucial for LL rhythmicity (e.g., LNd and DN1). Additionally, or alternatively, the manipulated l-LNv could generate signals interfering with normal network function, resulting in the observed rhythmic LL behavior.
Although qsm is a clock-controlled gene (16, 28, 46), the acute blue-light effects we observed are too fast to be mediated by transcriptional changes. Therefore, we favor a more direct membrane-localized mechanism in which rapid light-dependent posttranslational changes of Qsm alter the activity of Shaw and NKCC. Because (i) Cry is required for light-dependent Tim degradation in l-LNv (15), (ii) changing the Qsm level has no effect on Cry levels, and (iii) qsmOX triggers Tim degradation in the absence of Cry (16), the most likely explanation for the results reported here is that, in addition to activating Hk, Cry acts upstream of Qsm, which in turn regulates the activity of Shaw and NKCC. We assume that Qsm is activated by light because a light pulse at night rapidly increases protein levels (16). Qsm is an extracellular zona-pellucida (ZP) membrane-anchored protein, and we hypothesize that after light exposure the extracellular ZP domain is cleaved at a conserved furin protease cleavage site, a form of posttranslational processing typical for ZP-domain proteins (47). It is also possible that Qsm signals to Shaw and NKCC in both membrane-bound and cleaved forms. For example, at night membrane-bound Qsm could block NKCC, whereas light-induced cleavage could release this block, and the freed extracellular part could inactivate Shaw (Fig. 5B). This mechanism is reminiscent to the mechanism by which the GPI-anchored extracellular protein Sleepless increases Shaker (KV1 channel) activity for regulating Drosophila sleep (48).
How Qsm-induced changes in clock neuron activity influence the molecular clock remains an open question. Recent work shows that, in addition to the canonical degradation via Cry and Jetlag (49), Tim is also degraded via a Cul-3– and neuronal activity-dependent pathway in DD that has been implicated in mediating phase delays in the circadian clock (50, 51). In contrast to this activity-dependent Cul-3 pathway, the light responses in the current study depend on Cry. We therefore favor a model in which the combined functions of Qsm, Shaw, and NKCC contribute to the canonical Cry- and Jetlag-dependent Tim-degradation pathway.
In conclusion, we demonstrate that Qsm affects both daily and acute light responses of l-LNvs, and therefore (Qsm) presumably contributes to light-input to the Drosophila circadian clock. Qsm possibly signals downstream of Cry and acts on Shaw and NKCC to change clock neuronal activity in response to light.
Materials and Methods
Flies were obtained from fly stock centers or were generated using the ΦC31 integrase system (52) and were raised under standard conditions. The efficiency of RNAi knockdown was confirmed using quantitative RT-PCR (RT-qPCR) and a standard RNeasy kit (Qiagen). Analysis of fly activity was performed using the Drosophila Activity Monitor (DAM) System and either a MatLab tool-box (53) or ActogramJ (54). Electrophysiological recordings were performed as previously described (55). Prism (GraphPad) was used for statistical tests. Detailed procedures can be found in SI Materials and Methods.
SI Materials and Methods
Animals.
Flies were raised with a 12-h:12-h LD cycle on standard Drosophila medium (0.7% agar, 1.0% soy flour, 8.0% polenta/maize, 1.8% yeast, 8.0% malt extract, 4.0% molasses, 0.8% propionic acid, 2.3% nipagen) at 25 °C and were collected ∼3–5 d post eclosion. The following flies used in this study were described previously or obtained from the Bloomington, Vienna, or NIG fly stock center: gl60j (15), norpAP41 (37), HdcJK910 (38), and cry02 (56). For wild-type control recordings either Pdf-RFP (8) or Pdf-gal4 (57) flies crossed to uas-mCD8-RFP (BL27392) or uas-mCherry (BL52268) flies were used. For the behavioral controls wild-type (y w) and tim-gal4:16 (58) flies were used. Experimental genotypes including uas-qsm#5 (qsmOX) (16), uas-qsmRNAi (VDRC15394) and uas-qsmRNAi(2) (qsmRNAi) (16), uas-ShawWT12B (ShawOX) (21), uas-ShawRNAi (ShawRNAi) (21), and uas-CG31547RNAi (NKCCRNAi, NIG2509R-2) were crossed to tim-gal4:16 for behavioral tests and to either Pdf-gal4; uas-mCD8-RFP or Pdf-gal4; uas-mCherry flies for electrophysiological recordings. qsm105, a gal4 line inserted in the qsm first intron that reduces qsm expression (16), was used to test for behavioral interactions of qsm with Shaw and NKCC. For respective tests of physiological interaction, Pdf-RFP; tim-gal4:16 uas-qsmRNAi(2) flies were crossed to either uas-mCD8-GFP (BL30001) or uas-ShawWT12B and uas-CG31547RNAi flies. Note that in both sets of electrophysiology experiments (i.e., those involving Pdf-gal4 and tim-gal4), the ratio of gal4 to uas constructs was always 1:2. To test for Cry-dependent effects, Pdf-RFP was crossed into the cry02 background (56). The light sensitivity of wild-type flies is influenced by the naturally occurring s-tim/ls-tim polymorphism (35, 36). ls-tim flies are less light sensitive than s-tim flies, and in conjunction with jetlag mutants ls-tim elicits LL rhythmicity (35, 36). Therefore all fly stocks used in this study were genotyped for timeless and jetlag polymorphisms as described (35, 36) and, if necessary, were crossed into the ls-tim background to allow comparison between genotypes (all stocks were jetlag+). To reveal the effect of the s/ls-tim polymorphism, selected genotypes were also analyzed in an s-tim background (Table S1).
Generation of Transgenic Flies.
A flag-HA–tagged uas construct of CG31547-PB (UFO03679) was obtained from the Drosophila Genetics Resource Center (59) (www.fruitfly.org/EST/proteomics.shtml). uas-CG31547-flgHA was integrated into the ZH-attP-86Fb landing site using the ΦC31 integrase system to generate NKCCOX (52). The eye-expressed 3xP3-RFP cassette present in the landing site was eliminated by Cre-mediated excision as described (52).
RNA Isolation and RT-qPCR.
To test the efficiency of NKCCRNAi, 5- to 10-d-old flies were frozen at ZT 2, and 20 heads of each genotype were collected over dry ice. The total RNA was extracted using an RNeasy kit (QIAGEN) according to the manufacturer’s instructions and was finally eluted in RNase-free water and stored at −80 °C. cDNA synthesis was performed with the Reverse Transcription Reagents Kit (Applied Biosystems) in 20-μL reactions using 1 μg of total RNA. To verify the level of NKCC mRNA expression, dilutions of cDNA were used for PCR with rp49 primers, followed by DNA electrophoresis on 2% agarose gels to visualize the PCR products. Taqman probes for NKCC (catalog no. 4351372; Thermo Fisher) were applied to determine the amount of mRNA. Real-time assays were performed using an ABI GeneAMP PCR System 9700 using the standard program, and threshold cycle (CT) values were applied to determine the amount of RNA in each genotype. The relative concentrations were calculated using the comparative CT method, and RPL32 was used as control.
Behavior.
Analysis of the locomotor activity of 4- to 5-d-old male flies was performed using the Drosophila Activity Monitor System (DAM2; Trikinetics Inc.) with individual flies in recording tubes containing food (2% agar, 4% sucrose). The DAM monitors and an environmental monitor (DEnM; Trikinetics Inc.) were located inside a light- and temperature-controlled incubator (Percival Scientific Inc.) in which the fly’s activity was monitored for 4 d in a 12-h:12-h LD cycle followed by 7 d under LL (10 µW/cm2) at 25 °C. Plotting of behavioral activity, period calculations, and the determination of rhythmic statistics were performed using either a signal-processing tool-box (53) implemented in Matlab (MathWorks) or the ImageJ (https://imagej.nih.gov/ij/) plug-in ActogramJ (54). LL rhythmicity was determined on the basis of a rhythmic statistics value >1.5 and was classified according to the period length of individual flies as being either bimodal and circadian (∼12 h or ∼24 h, respectively) or ultradian (<10 h). The significance of rhythmicity for ultradian periods was determined using periodogram analysis in ActogramJ (P value cut-off set to 0.001); each period was subsequently verified by visual inspection of the actograms. For simplicity the value of the overall rhythmicity includes flies of all categories, but the average period was calculated based on the prevalent period for each genotype.
Electrophysiological Recordings.
Patch-clamp recordings were performed as described previously (55). Flies were decapitated, and brains were dissected either at ZT 1–3 (daytime condition) or ZT 13–15 (nighttime condition). For recordings at night, dissections were performed on a M205C stereomicroscope (Leica) set to minimal white light, and experiments were conducted under red-light illumination. Whole fly brains were acutely dissected in extracellular saline solution containing (in mM): 101 NaCl, 1 CaCl2, 4 MgCl2, 3 KCl, 5 glucose, 1.25 NaH2PO4, 20.7 NaHCO3, pH 7.2. Some brains were transferred briefly in saline containing 20 U/mL papain and 1 mM l-cysteine to aid in the removal of the ganglion sheath. After removal of the photoreceptors, lamina, air sacks, and trachea, a small incision was made over the position of the l-LNv neurons to give easier access for the recording electrodes. The brain then was placed ventral side up in the recording chamber, secured using a custom-made anchor, and during recordings was continuously perfused with aerated (95% O2, 5% CO2) saline solution. Neurons were visualized using a 63× lens and a 555-nm LED light source on an upright Zeiss microscope (Examiner.Z1; Carl Zeiss Microscopy GmbH). l-LNv neurons were identified on the basis of their fluorescence, size, and position (Fig. S3 A and B). Generally, a single recording from one l-LNv per brain was performed at room temperature (20–22 °C). If a recording was performed from a second neuron in the same brain, the neuron was on the opposite side of the brain, and the recording was made at least 20 min after the first recording. Whole-cell current- and voltage-clamp recordings were performed using glass electrodes with 8- to 18-MΩ resistance filled with intracellular solution (in mM: 102 K-gluconate, 17 NaCl, 0.94 EGTA, 8.5 Hepes, 0.085 CaCl2, 1.7 MgCl2 or 4 Mg⋅ATP, and 0.5 Na·GTP, pH 7.2) and an Axon MultiClamp 700B amplifier, digitized with an Axon DigiData 1440A (sampling rate: 20 kHz; filter: Bessel 10 kHz), and recorded using pClamp 10 (Molecular Devices). Correction was made for capacitance, series resistance, and leak currents, and voltage-clamp traces were low-pass filtered offline (100-Hz cutoff; 8-Pole Bessel). All chemicals were purchased from Sigma.
Light and GABA Application.
To test the effect of light on the activity of the l-LNvs, a blue-light pulse (470 nm) was applied using the fluorescence kit and appropriate filter of the microscope (Colibri; Zeiss) set at 50% intensity (7.3 mW/cm2). As controls, other wavelengths (green, 555 nm, and red, 625 nm) were also tested. The experimental design consisted of 1 min of recording before light application, the 30-s light exposure, and further recording for 2.5 min afterwards. To test for changes in the EGABA, GABA (25 mM) was injected (10-ms pulse) in the ipsilateral medulla via a glass pipette (1–3 MΩ) and a Picospritzer III (10 psi; Parker Hannifin).
Data Analysis/Statistics.
The liquid junction potential of the recordings was calculated as 13 mV and was subtracted from all the membrane voltages. The RMP was measured after stabilization for 2–3 min after achieving whole-cell configuration. The SFR and spike frequency (F) were measured using 10-s bins throughout the experiment. To quantify the effect of blue light on the firing rate, the average frequency of the 30-s baseline before light application was subtracted from the average of the 30-s light-on frequency (Fon − Foff). A cell was included in the analysis if the access resistance was less than 50 MΩ, and only tonically firing neurons were analyzed (a total of three bursting neurons were rejected). EGABAs were calculated from the I–V curves resulting from the currents elicited by pressure-injected GABA at different holding potentials in the voltage-clamp mode. Sustained K+ currents were measured from the average of the last 100 ms of a 1-s voltage pulse obtained from neurons held at a potential of −80 mV and stimulated to potentials up to +100 mV in 10-mV intervals. The current density was calculated by normalizing the measured currents with the cell capacitance. Because overexpression and RNAi knockdown of qsm, Shaw, and NKCC eliminated the day/night differences in physiological parameters, recordings to measure the EGABA and the K+ currents of l-LNvs with altered Qsm, Shaw, or NKCC levels were performed at both daytime and nighttime. Again, we did not observe a difference between daytime and nighttime recordings, and therefore the data were pooled. All values are given as mean and SD. The Kolmogorov–Smirnov test was used to test for normality; the t test [indicated by a hash mark (#) in the figures] was used to calculate significant differences between wild-type control flies at day and at night for RMP, SFR, and Fon − Foff; a one-way ANOVA followed by Dunnett’s multiple comparison test was used for differences between wild-type and test flies within the day or night condition for RMP, SFR, and Fon-Foff or followed by Tukey’s multiple comparison test for differences in the EGABA and KV current densities (indicated in the figures by an asterisk). The statistical tests were performed using Prism (GraphPad Software Inc.), and the figures were arranged in Illustrator (Adobe Systems Inc.).
Acknowledgments
We thank Min Xu for generating the uas-qsmOX and uas-NKCCOX transgenics, Drs. Alan Roberts and Maria Usowicz for advice and helpful comments on the manuscript, and the Joerg Albert laboratory for assistance and equipment to perform the qPCR. This work was supported by Biotechnology and Biological Sciences Research Council Grants BB/J017221/1 (to J.J.L.H.) and BB/J018589/2, (to R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606547113/-/DCSupplemental.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- 2.Helfrich-Förster C, Nitabach MN, Holmes TC. Insect circadian clock outputs. Essays Biochem. 2011;49(1):87–101. doi: 10.1042/bse0490087. [DOI] [PubMed] [Google Scholar]
- 3.Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15(17):R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417(6886):329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 5.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28(25):6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheeba V, et al. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18(20):1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizrak D, et al. Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons. Curr Biol. 2012;22(20):1871–1880. doi: 10.1016/j.cub.2012.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruben M, Drapeau MD, Mizrak D, Blau J. A mechanism for circadian control of pacemaker neuron excitability. J Biol Rhythms. 2012;27(5):353–364. doi: 10.1177/0748730412455918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci. 2011;12(10):553–569. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109(4):485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 12.Meijer JH, Michel S. Neurophysiological analysis of the suprachiasmatic nucleus: A challenge at multiple levels. Methods Enzymol. 2015;552:75–102. doi: 10.1016/bs.mie.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95(5):669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 14.Stanewsky R, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95(5):681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 15.Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30(1):249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 16.Chen KF, Peschel N, Zavodska R, Sehadova H, Stanewsky R. QUASIMODO, a novel GPI-anchored zona pellucida protein involved in light input to the Drosophila circadian clock. Curr Biol. 2011;21(9):719–729. doi: 10.1016/j.cub.2011.03.049. [DOI] [PubMed] [Google Scholar]
- 17.Yoshii T, et al. Cryptochrome-dependent and -independent circadian entrainment circuits in Drosophila. J Neurosci. 2015;35(15):6131–6141. doi: 10.1523/JNEUROSCI.0070-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331(6023):1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fogle KJ, et al. CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel β-subunit redox sensor. Proc Natl Acad Sci USA. 2015;112(7):2245–2250. doi: 10.1073/pnas.1416586112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lear BC, et al. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron. 2005;48(6):965–976. doi: 10.1016/j.neuron.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Hodge JJL, Stanewsky R. Function of the Shaw potassium channel within the Drosophila circadian clock. PLoS One. 2008;3(5):e2274. doi: 10.1371/journal.pone.0002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flourakis M, et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell. 2015;162(4):836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itri JN, Michel S, Vansteensel MJ, Meijer JH, Colwell CS. Fast delayed rectifier potassium current is required for circadian neural activity. Nat Neurosci. 2005;8(5):650–656. doi: 10.1038/nn1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamble KL, Kudo T, Colwell CS, McMahon DG. Gastrin-releasing peptide modulates fast delayed rectifier potassium current in Per1-expressing SCN neurons. J Biol Rhythms. 2011;26(2):99–106. doi: 10.1177/0748730410396678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo T, Loh DH, Kuljis D, Constance C, Colwell CS. Fast delayed rectifier potassium current: Critical for input and output of the circadian system. J Neurosci. 2011;31(8):2746–2755. doi: 10.1523/JNEUROSCI.5792-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alamilla J, Perez-Burgos A, Quinto D, Aguilar-Roblero R. Circadian modulation of the Cl(-) equilibrium potential in the rat suprachiasmatic nuclei. BioMed Res Int. 2014;2014:424982. doi: 10.1155/2014/424982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones JR, Tackenberg MC, McMahon DG. Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat Neurosci. 2015;18(3):373–375. doi: 10.1038/nn.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kula-Eversole E, et al. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci USA. 2010;107(30):13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parisky KM, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60(4):672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA. 2008;105(50):19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2008;99(2):976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508(6):952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 33.Emery P, Stanewsky R, Hall JC, Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000;404(6777):456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- 34.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302(5651):1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 35.Peschel N, Veleri S, Stanewsky R. Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila’s circadian clock. Proc Natl Acad Sci USA. 2006;103(46):17313–17318. doi: 10.1073/pnas.0606675103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandrelli F, et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science. 2007;316(5833):1898–1900. doi: 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- 37.Szular J, et al. Rhodopsin 5- and Rhodopsin 6-mediated clock synchronization in Drosophila melanogaster is independent of retinal phospholipase C-β signaling. J Biol Rhythms. 2012;27(1):25–36. doi: 10.1177/0748730411431673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieger D, Stanewsky R, Helfrich-Förster C. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J Biol Rhythms. 2003;18(5):377–391. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- 39.Wei A, et al. K+ current diversity is produced by an extended gene family conserved in Drosophila and mouse. Science. 1990;248(4955):599–603. doi: 10.1126/science.2333511. [DOI] [PubMed] [Google Scholar]
- 40.Salkoff L, et al. An essential ‘set’ of K+ channels conserved in flies, mice and humans. Trends Neurosci. 1992;15(5):161–166. doi: 10.1016/0166-2236(92)90165-5. [DOI] [PubMed] [Google Scholar]
- 41.Murad A, Emery-Le M, Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron. 2007;53(5):689–701. doi: 10.1016/j.neuron.2007.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 2007;5(11):e315. doi: 10.1371/journal.pbio.0050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoleru D, et al. The Drosophila circadian network is a seasonal timer. Cell. 2007;129(1):207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 44.Hodge JJL, Choi JC, O’Kane CJ, Griffith LC. Shaw potassium channel genes in Drosophila. J Neurobiol. 2005;63(3):235–254. doi: 10.1002/neu.20126. [DOI] [PubMed] [Google Scholar]
- 45.Hebert SC, Mount DB, Gamba G. Molecular physiology of cation-coupled Cl- cotransport: The SLC12 family. Pflugers Arch. 2004;447(5):580–593. doi: 10.1007/s00424-003-1066-3. [DOI] [PubMed] [Google Scholar]
- 46.Stempfl T, et al. Identification of circadian-clock-regulated enhancers and genes of Drosophila melanogaster by transposon mobilization and luciferase reporting of cyclical gene expression. Genetics. 2002;160(2):571–593. doi: 10.1093/genetics/160.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plaza S, Chanut-Delalande H, Fernandes I, Wassarman PM, Payre F. From A to Z: Apical structures and zona pellucida-domain proteins. Trends Cell Biol. 2010;20(9):524–532. doi: 10.1016/j.tcb.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Wu MN, et al. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13(1):69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19(3):241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 50.Grima B, Dognon A, Lamouroux A, Chélot E, Rouyer F. CULLIN-3 controls TIMELESS oscillations in the Drosophila circadian clock. PLoS Biol. 2012;10(8):e1001367. doi: 10.1371/journal.pbio.1001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. eLife. 2014;3:e02780. doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104(9):3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3(1):1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmid B, Helfrich-Förster C, Yoshii T. A new ImageJ plug-in “ActogramJ” for chronobiological analyses. J Biol Rhythms. 2011;26(5):464–467. doi: 10.1177/0748730411414264. [DOI] [PubMed] [Google Scholar]
- 55.Chen C, et al. Drosophila Ionotropic Receptor 25a mediates circadian clock resetting by temperature. Nature. 2015;527(7579):516–520. doi: 10.1038/nature16148. [DOI] [PubMed] [Google Scholar]
- 56.Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics. 2007;177(1):329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park JH, Hall JC. Isolation and chronobiological analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster. J Biol Rhythms. 1998;13(3):219–228. doi: 10.1177/074873098129000066. [DOI] [PubMed] [Google Scholar]
- 58.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422(1):66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 59.Yu C, et al. 2011. Development of expression-ready constructs for generation of proteomic libraries. Protein Microarray for Disease Analysis, Methods in Molecular Biology, ed Wu CJ (Humana, Valley Stream, NY), Vol 723, pp 257–272.