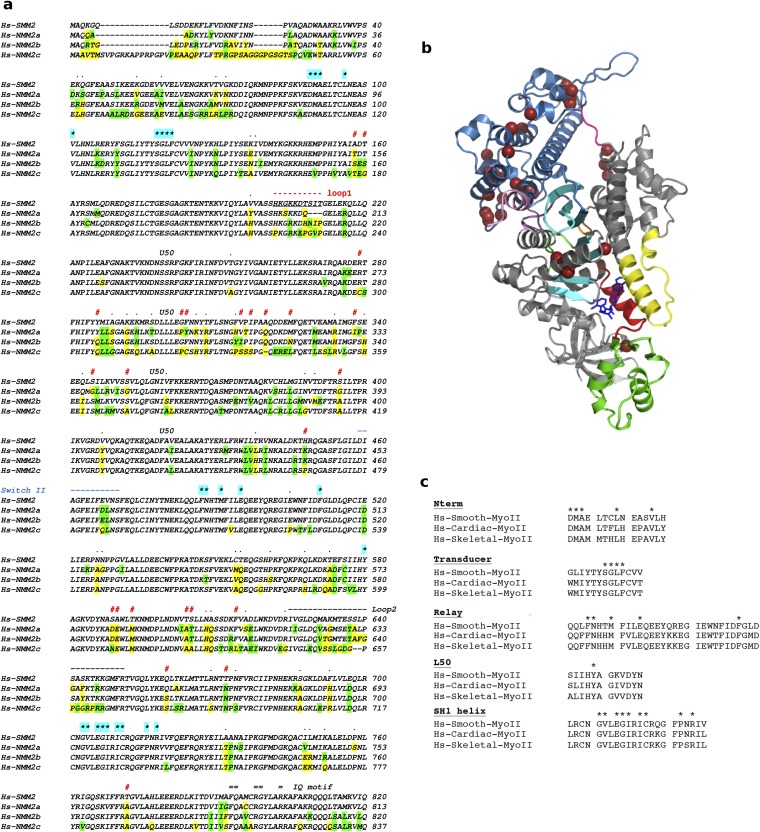
Fig. S1.
(A) Sequence alignment of the heavy chain of Hs-SMM2 and Hs-NMM2a, -b, and -c. Residues that are not conserved are shown in yellow; similar residues that are not conserved are green. Residues involved in the binding of CK-571 are labeled with a star highlighted cyan: none of these residues are different in sequence between SMMs and NMMs. Human SMM2 and NMM2a, NMM2b, and NMM2c share, respectively, 83%, 84%, and 75% sequence identity and 91%, 91%, and 88% sequence similarity in their MD. Note the main sequence differences noted with the symbol # and the sequence for loop1 and switch II indicated, respectively, in red and blue. In between these two elements, the sequence corresponds to residues of the U50 subdomain. The kinetics of the allosteric rearrangements occurring in the recovery stroke depends on the full sequence of the myosin motor and determines the specific kinetic rates of ATP hydrolysis. In transient kinetic studies, the rate that measures both the recovery stroke isomerization and hydrolysis (k+3 + k-3) is faster for SMM2 (43) (k+3 + k-3 = 50 s−1) compared with NMM2A (44) (k+3 + k-3 = 14 s−1) and NMM2B (45) (k+3 + k-3 = 16.7 s−1). Differences in the U50 subdomain or the loop1 sequence are the most likely candidates to influence the overall dynamics in this transition, in particular by controlling the rearrangements necessary in the transducer for this transition (Fig. S4C). Thus, specificity in the inhibition depends on the dynamics of this reversible transition that is modulated by residues that are far from the drug binding site. (B) Residues that are not conserved between Hs-SMM2 and NMM2 and are likely candidates to modulate the recovery stroke are shown with brown spheres on the SMM/CK-571 structure (see also Movie S4). (C) Sequence alignment of Human Skeletal, Cardiac, and Smooth Myosin II. Residues involved in CK-571 binding are labeled with a star. Note that they are all identical in these different myosins.