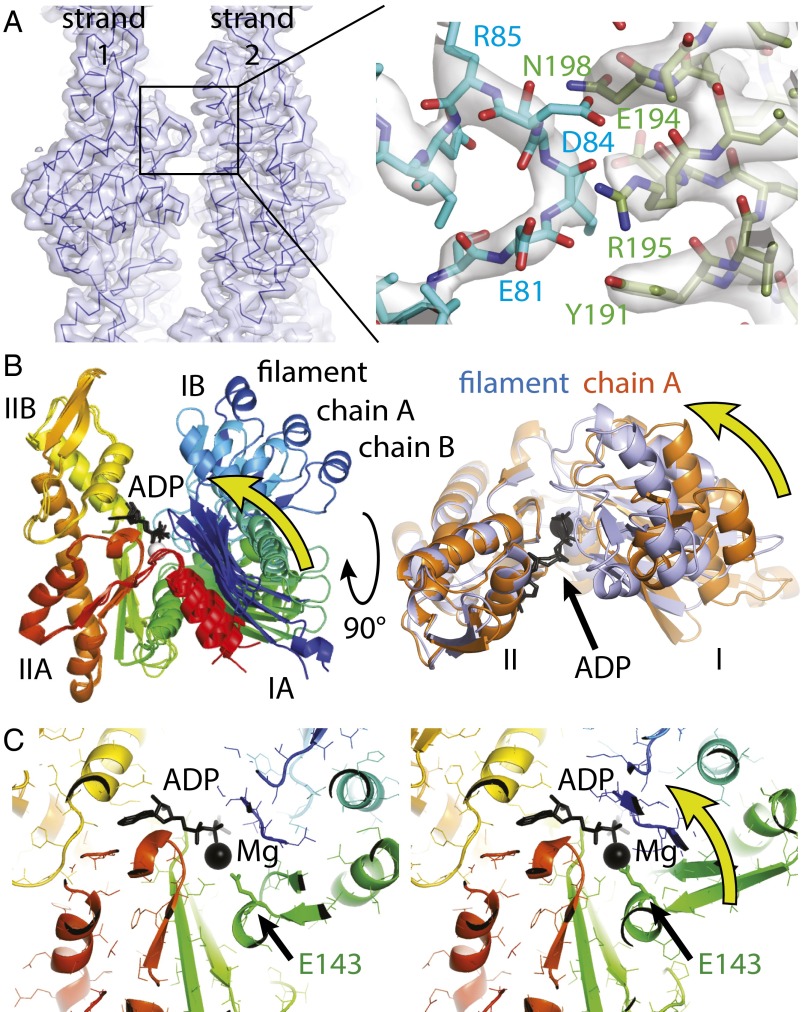
Fig. 3.
MamK lateral contacts and conformational changes upon polymerization. (A) The two protofilaments (strands) generating the double-helical MamK filament are held together by a single contact. The contact (detailed Right) is formed by two consecutive stretches of residues, 81 to 84 on one side and 191 to 198 on the other. Because this contact repeats exactly on the other side (through the C2 symmetry of the filament) and also along the filament between each pair of subunits, avidity keeps the protofilaments together despite such a small contact area. (B) The structure of MamK in the filament closes further, rotating domains I and II against each other by 19.8° such that the cleft between subdomains IB and IIB becomes smaller and also removing a propeller twist between domains I and II, as determined by DynDom (27). (B, Right) Removal of the propeller twist causes several loops (orange to purple) to close around the nucleotide (black). Directions of movements are indicated by yellow arrows. All structural alignments were performed superimposing domain II only (residues 156 to 314). See also Movies S3 and S4. (C) The conformational change upon polymerization moves a key residue, Glu-143, closer to the magnesium ion, presumably switching ATP hydrolysis during the polymerization cycle of MamK. As in B, the direction of the movement is indicated by a yellow arrow. See also Movie S5.