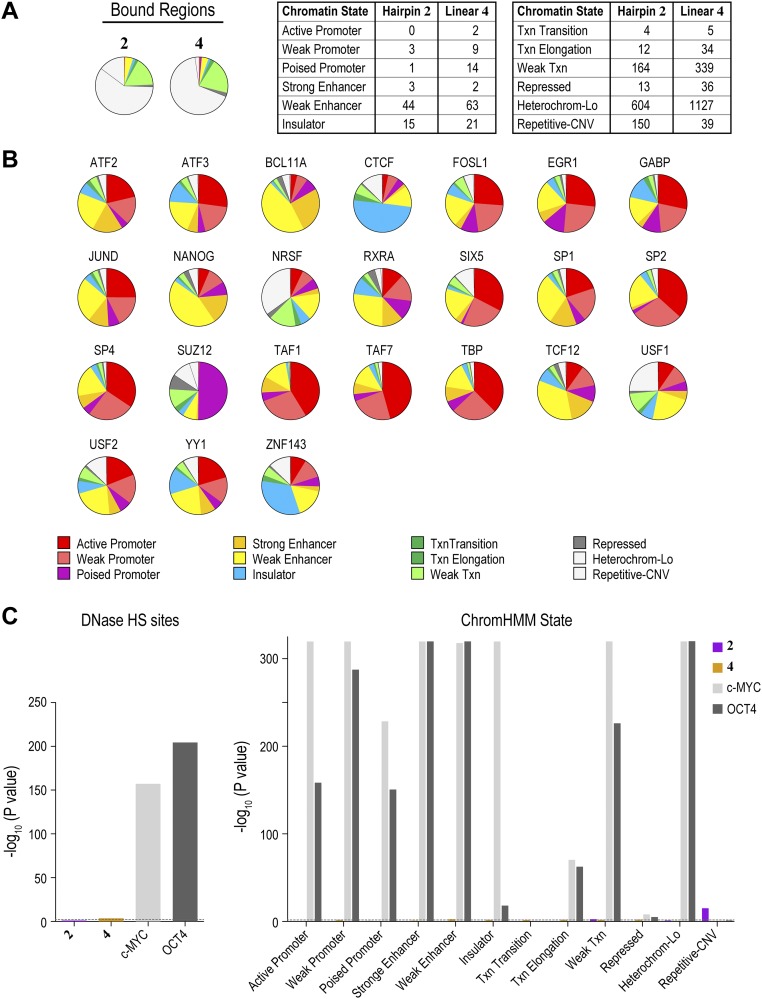
Fig. S12.
Synthetic genome readers distribute across diverse chromatin states. (A) Polyamide derivatives 2 and 4 show distributions across chromatin states that resemble the distributions predicted by CSI-Genomescapes. Distribution of loci predicted to be bound by 2 and 4 across the 12 chromatin states. For reference, the distribution of regions observed by COSMIC-seq is shown. (B) Distribution of 24 endogenous transcription factors across the 12 chromatin states in hESCs (32). (C) Two-tailed Fisher’s exact test confirms that endogenous transcription factors exhibit strong preferences for specific DNase HS sites and chromatin states, whereas 2 and 4 show little preference for DNase HS sites or specific chromatin states. The dashed line indicates a Bonferroni-adjusted P value of 0.0038.