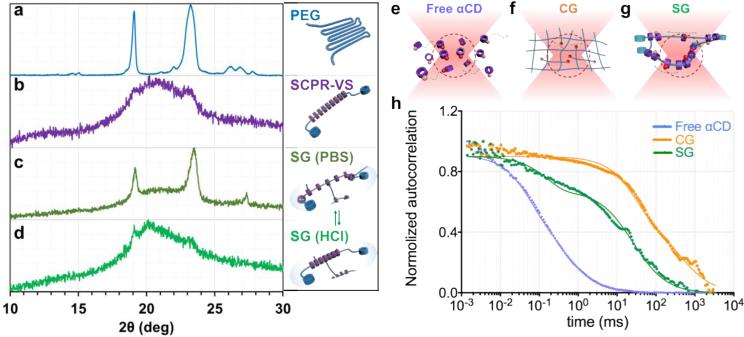
Figure 2. Sliding hydrogels (SG), but not chemical hydrogels (CG), demonstrate reversible ligand distribution and dynamic molecular ligand mobility, driven by sliding αCDs along the PEG backbone in 3D.
(a-d) X-ray diffraction (XRD) pattern confirms reversible distribution of αCDs in sliding hydrogel. a: Baseline pattern of PEG backbone without αCD; b: Crystal structure formation confirms packing of sliding CDs along PEG backbone; c. Sliding hydrogels in PBS; αCDs are ionized in crosslinked SG, reverting from packed pattern to more dispersed distribution, exposing structure of PEG backbone similar to a; d. Upon switching to HCL solution, αCDs in sliding hydrogels are deionized, reverting to packed state similar to pattern in b. (e-g) Scheme of movement of tetramethylrhodamine-labeled ligands (red spheres) toward and away from the focal volume (red dotted region) of confocal microscopy when they are tethered on (e) free αCDs, (f) chemical hydrogel (CG), and (g) sliding hydrogel (SG). (h) Normalized autocorrelation curve from fluorescence correlation spectroscopy showed fastest decay of free αCDs (blue) due to their free diffusion. The sliding hydrogel (SG; green) exhibited a faster decay of auto-correlation than the chemical hydrogel (CG; orange), indicating higher mobility, but more stable than freely diffusive αCDs.