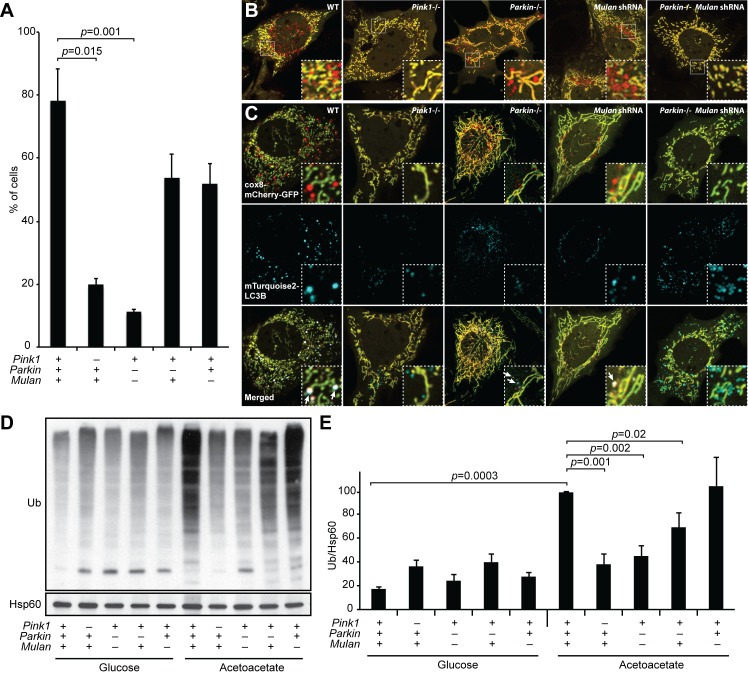
Figure 4. MUL1 and PARKIN have redundant functions in OXPHOS-induced mitophagy.
(A) Quantification of red-only puncta in cells grown in acetoacetate-containing medium. Presence (+) or absence (-) of Pink1, Parkin, or Mul1 is indicated. Error bars indicate SD of three biological replicates, p=0.015 (Pink1), p=0.0011 (Parkin-/- Mulan shRNA) (Student’s t-test). (B) Mitophagy in wild-type and mutant cells. Cells stably expressing Cox8-EGFP-mCherry were grown in acetoacetate-containing medium and imaged by fluorescence microscopy. (C) Co-localization of LC3B with mitophagy intermediates. Wild-type and mutant cells were retrovirally transduced with mTurquoise2-LC3B, grown in acetoacetate-containing medium and imaged by fluorescence microscopy. Examples of LC3B co-localization with mitophagy intermediates are indicated by arrows. (D) Accumulation of polyubiquitinated proteins in mitochondria. Cells were grown in the indicated medium, and mitochondria were isolated by differential centrifugation. Mitochondrial lysates were analyzed by Western blot for pan-Ubiquitin. HSP60 is a loading control. (E) Quantification of polyubiquitinated proteins in mitochondria. Three independent experiments were quantified by densitometry and averages are shown. Ubiquitin level was normalized to HSP60. Error bars indicate SD, p=0.0003 (WT Glu vs. Ac), p=0.0011 (Pink1-/-), p=0.0016 (Parkin-/- Mulan shRNA), p=0.0206 (Parkin-/-) (Student’s t-test).