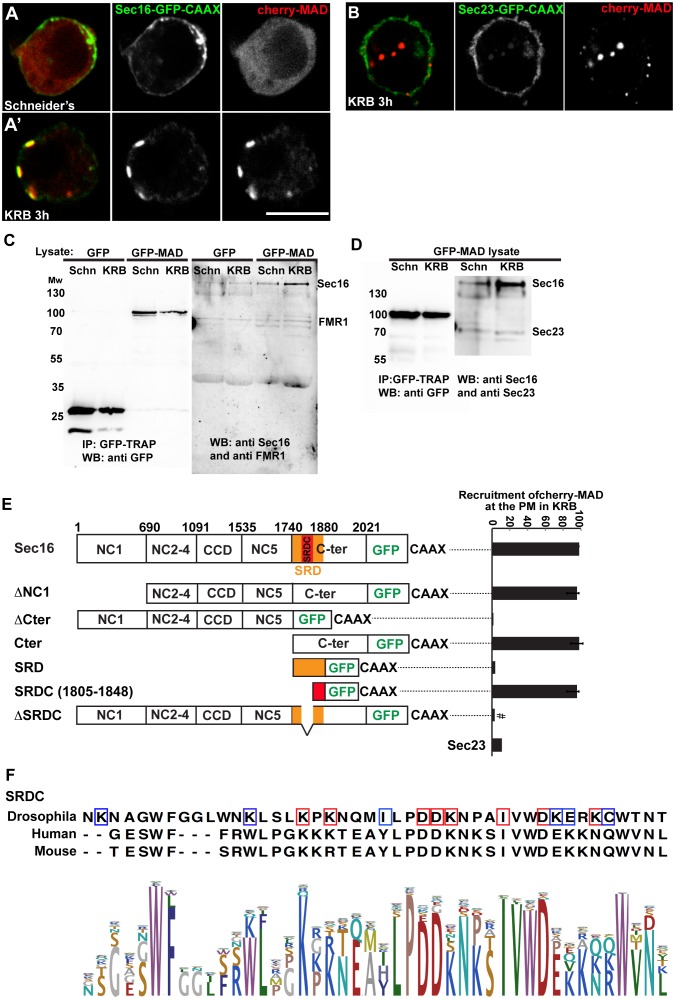
Figure 7. Sec16 SRCD is MARylated.
(A, A’) Co-visualisation of full length Sec16-GFP-CAAX and Cherry-MAD in growing (Schneider’s) S2 cells (A) and upon amino-acid starvation (KRB) (A’). Note that Sec16-GFP-CAAX localises to the plasma membrane where cherry-MAD is recruited upon amino-acid starvation (KRB), whereas in Schneider’s, it remains cytoplasmic. (B) Co-visualisation of full length Sec23-GFP-CAAX (B) and cherry-MAD upon amino-acid starvation (KRB). Note that cherry-MAD is not recruited to the plasma membrane and forms spots in the cytoplasm. (C) WB (using anti GFP, anti Sec16, anti FMR1) of GFP-MAD immuno-precipitation (IP) using GFP-TRAP from stable S2 cell lines expressing GFP-MAD and GFP, either in growing conditions (Schneider’s) or upon amino-acid starvation (KRB 3 hr). (D) WB (using anti GFP, anti Sec16, anti Sec23) of GFP-MAD IP from stable S2 cells expressing GFP-MAD, either in growing conditions (Schneider’s) or upon amino-acid starvation (KRB 3 hr). Note that Sec23 pull-down by GFP-MAD is very weak when compared to Sec16. (E) Map of all Sec16-GFP-CAAX truncations used and the quantitation of cherry-MAD recruitment to the plasma membrane. Note that Sec16-ΔSRDC-GFP-CAAX transfection was performed in Sec16 depleted cells (marked by #) to avoid oligomerisation with endogenous Sec16. (F) Muscle sequence alignment of Drosophila, human and mouse Sec16 SRDC (1805–1848) and presentation of logo sequence outlining the degree of sequence conservation among all eukaryotes (defined using CLC Main Workbench 6.7.1 using the full length Sec16 sequence against all eukaryote sequences from the non-redundant protein database using standard settings). The red squares indicate the conserved residues that are potentially MARylated and the blue squares the non-conserved ones.
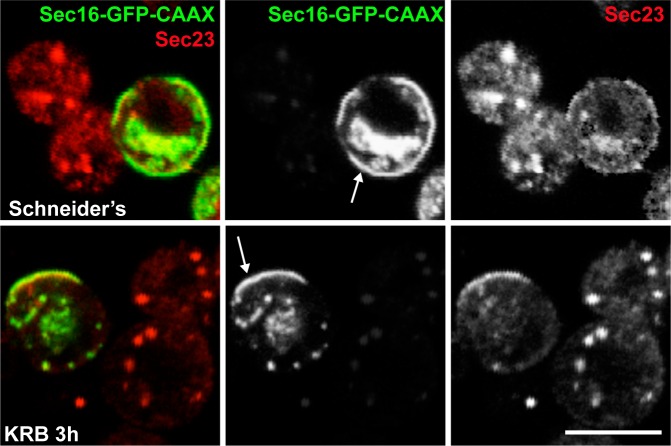
Figure 7—figure supplement 1. Sec16-CAAX recruits Sec23 to the PM upon AA starvation.
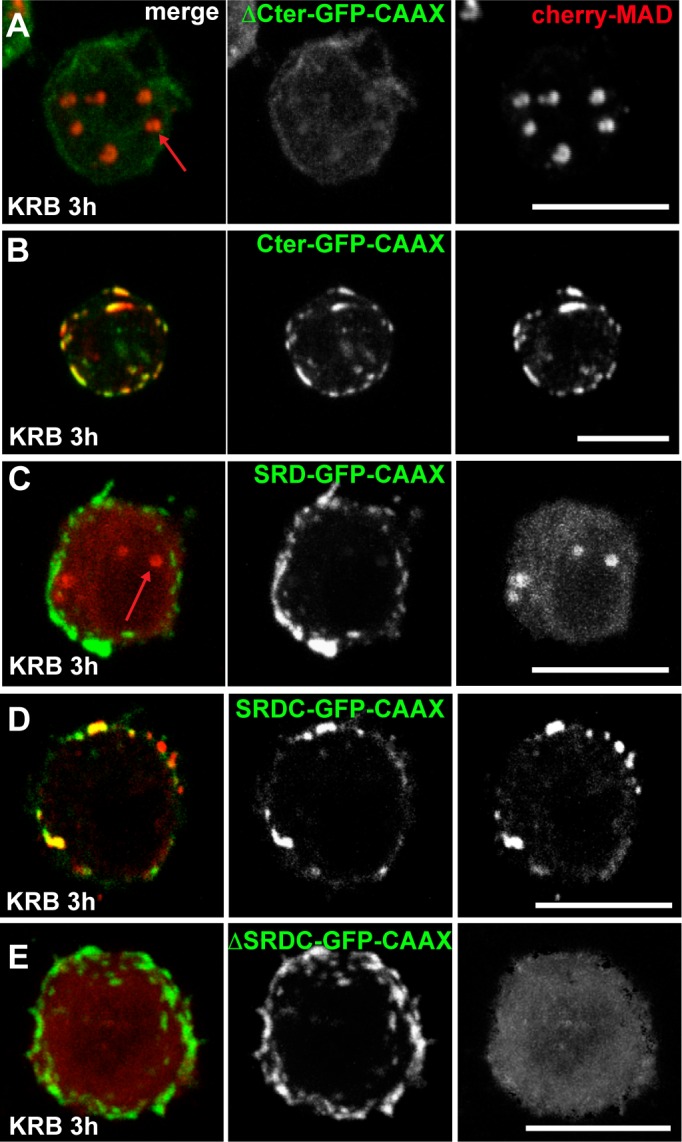
Figure 7—figure supplement 2. Sec16 SRDC is MARylated upon amino-acid starvation.