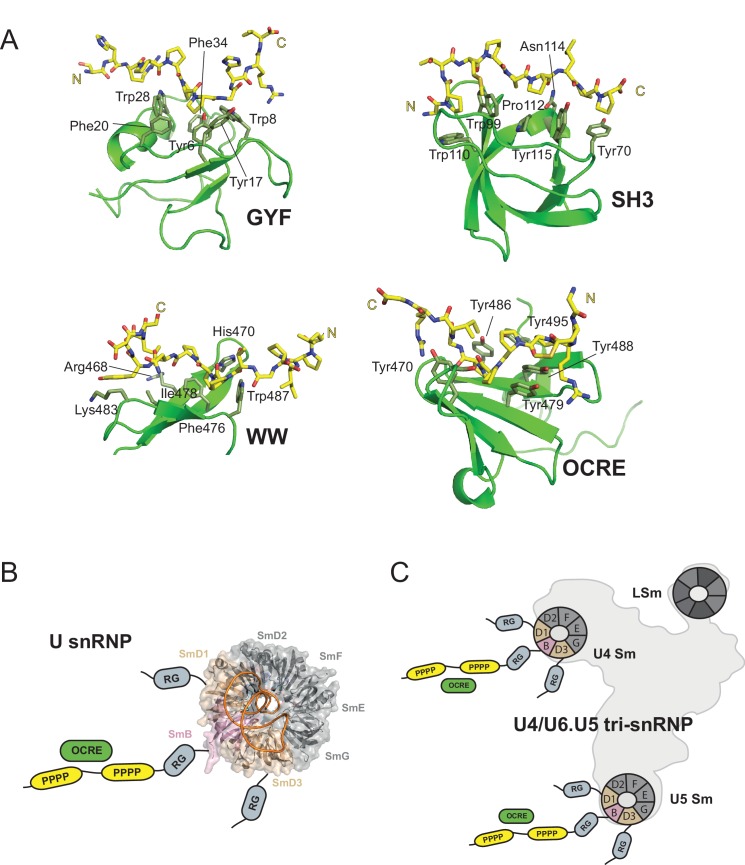
Figure 7. OCRE is a novel PRM-binding domain that interacts with snRNPs.
(A) Comparison of proline-rich motif (PRM) recognition by PRM binding domains.Cartoon representations of various PRM binding domains (green) with side chains of important residues shown as sticks and annotated. PRM ligands are shown by stick representation (yellow). PDB accession codes: GYF (PDB 1L2Z), SH3 (PDB accession 1ABO), WW (PDB 1I5H), and RBM5 OCRE. (B) The C-terminal tail of SmN/B/B’ comprising RG-rich regions and proline-rich motifs (PRM) extends far from the core Sm core fold in the seven-membered Sm ring of assembled U snRNPs. The crystal structure of U4 snRNP core (PDB 4WZJ) (Leung et al., 2011) is shown, the C-terminal tails of SmD1/D3, which harbour RG-rich regions and SmN/B/B’ comprising RG and PRM regions are not visible in the crystal structure and indicated schematically. (C) The seven-membered Sm core rings of U4 and U5 snRNPs are located at the outside of the assembled U4/U6.U5 tri-snRNP (Nguyen et al., 2015). Thus, also in the tri snRNP the SmN/B/B’ tails are accessible for interactions with the RBM5 OCRE domain.