Figure 3. Identification of retrotransposition-independent SLAVs.
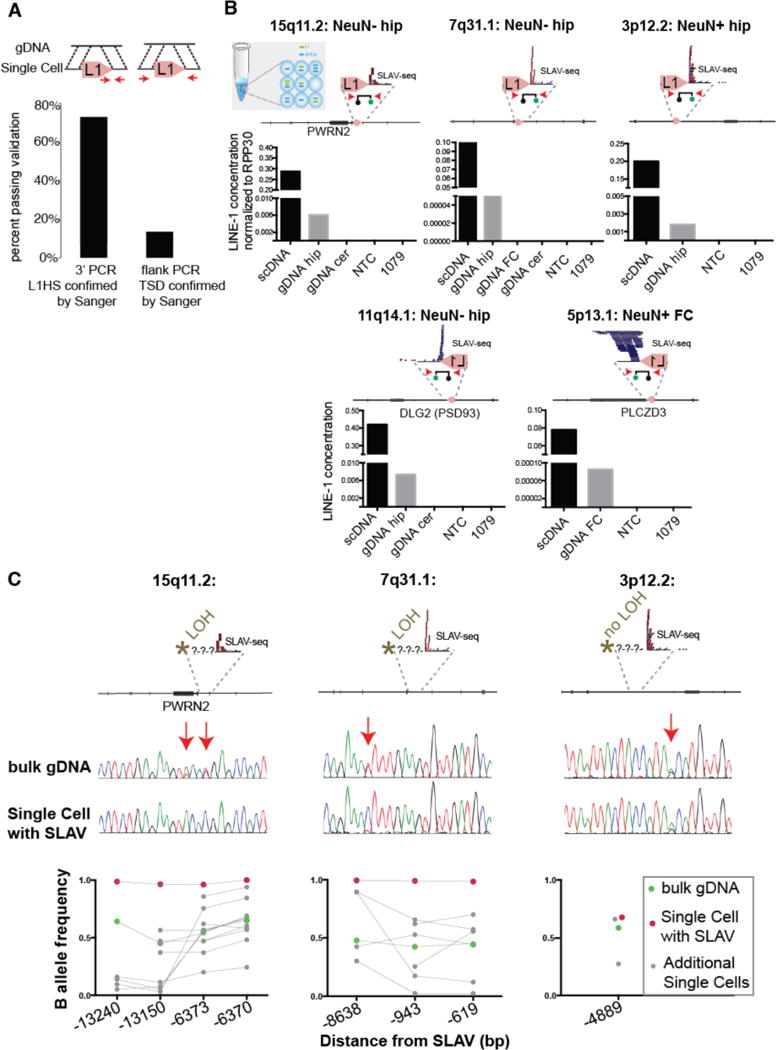
(A) Percentage of predicted variants validated by 3′ PCR-Sanger sequencing or flanking/TSD PCR-Sanger (arrows indicate PCR primers). (B) Digital PCR assay confirms the presence of the L1 variant in single cell (scDNA) and bulk genomic DNA (gDNA) (as described in Fig. 2C). The specified variants have a confirmed 3′-L1Hs junction but lack TSD. (C) Loss of heterozygosity is detected upstream of the variant in 2 of 3 specified variants. The arrows indicate single nucleotide polymorphisms detected in bulk gDNA. Bottom: Quantification of additional SNVs from single cell and bulk genomic DNA. Connecting line indicates the same DNA sample tested across several positions.
