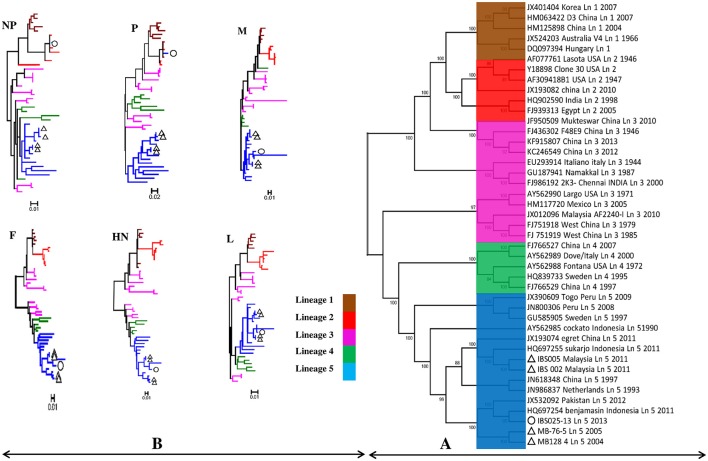
Figure 2.
The maximum likelihood phylogenetic trees constructed by using 5 Malaysian NDV isolates. (A) The phylogenetic tree based on nucleotide sequence of complete genome compared with 38 representative complete genomes of lineage 1–5 belongs to APMV-1. (B) The phylogenetic trees based on amino acids sequence of NP, P, M, F, HN, and L proteins compared with 38 representative protein sequence of lineage 1–5 belongs to APMV-1. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA6. Malaysian isolates MB128/04, MB076/05, IBS002/11, and IBS005/11 presented in this study are marked with a triangle (Δ) and IBS025/13 with a circle (◦).