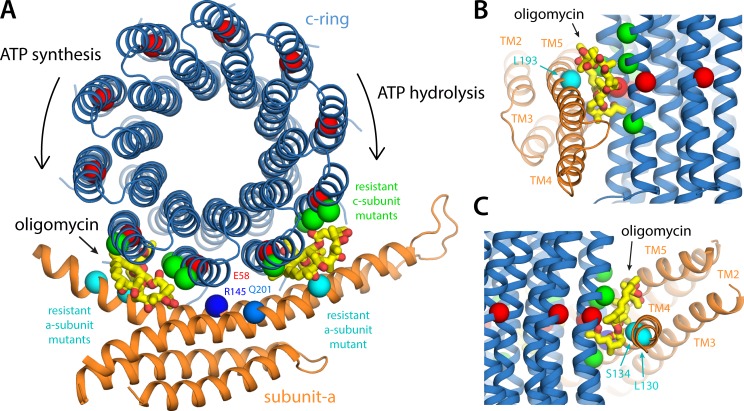
Figure 9.
Insights into the mechanism of bi-directional inhibition of the ATP synthase by oligomycin. (A) Structure of the a–c complex, viewed as in Fig. 6 A, with two oligomycin molecules (yellow/red) bound to the c-ring. The binding pose of oligomycin is exactly that observed in a cocrystal structure of the c-ring from S. cerevisiae (in the absence of subunit a; see Results and discussion). This pose appears to be entirely feasible with two different sites at the a–c interface, to the left and to the right of the site where Glu58 interacts with Arg145 (i.e., the P site). An oligomycin molecule would not fit in this P site, implying that binding at the flanking sites shown would ultimately halt the rotation of the ring. The positions in the S. cerevisiae c and a subunits where mutations confer resistance to oligomycin inhibition (see Results and discussion) are marked with green and cyan spheres, respectively. These positions are Leu52, Ala55, Leu56, and Phe63 in the outer helix of the c subunit (Leu53, Ala56, Leu57, and Phe64 in S. cerevisiae), Leu130 and Ser134 in aTM4, and Leu193 in aTM5 (aILe171, aSer175, and aLeu232 in S. cerevisiae). (B) Side view of the oligomycin inhibition site when the c-ring rotates clockwise, transporting H+ uphill, driven by ATP hydrolysis. (C) Side view of the inhibition site when the c-ring rotates counterclockwise, driven by downhill H+ permeation, during ATP synthesis.