Abstract
Background:
The purpose of this study was to evaluate single-nucleotide polymorphisms (SNPs) in genes encoding key metabolising enzymes or involved in pharmacodynamics for possible associations with paclitaxel-induced peripheral neuropathy.
Methods:
The study population consists of 188 women from the multicenter, randomised, phase II ATX trial (BOOG2006-06; EudraCT number 2006-006058-83) that received paclitaxel and bevacizumab without or with capecitabine as first-line palliative therapy of HER2-negative metastatic breast cancer. Genotyping of CYP2C8*3 (c.416G>A), CYP3A4*22 (c.522-191C>T), TUBB2A (c.-101T>C), FGD4 (c.2044-236G>A) and EPHA5 (c.2895G>A) was performed by real-time PCR. Toxicity endpoints were cumulative dose (1) until first onset of grade ⩾1 peripheral neuropathy and (2) until first paclitaxel dose reduction from related toxicity (NCI-CTCAE version 3.0). SNPs were evaluated using the Kaplan–Meier method, the Gehan–Breslow–Wilcoxon test and the multivariate Cox regression analysis.
Results:
The rate of grade ⩾1 peripheral neuropathy was 67% (n=126). The rate of dose reduction was 46% (n=87). Age ⩾65 years was a risk factor for peripheral neuropathy (HR=1.87, P<0.008), but not for dose reduction. When adjusted for age, body surface area and total cumulative paclitaxel dose, CYP2C8*3 carriers had an increased risk of peripheral neuropathy (HR=1.59, P=0.045). FGD4 c.2044-236 A-allele carriers had an increased risk of paclitaxel dose reduction (HR per A-allele=1.38, P=0.036) when adjusted for total cumulative paclitaxel dose.
Conclusions:
These findings may point towards clinically useful indicators of early toxicity, but warrant further investigation.
Keywords: genetic polymorphisms, CYP2C8*3, FGD4, paclitaxel, peripheral neuropathy, breast cancer
Paclitaxel has become part of the standard chemotherapeutic armamentarium for a wide range of solid tumours. Treatment with paclitaxel, however, is associated with specific toxicities, among which is peripheral sensory and motor neuropathy. Paclitaxel-induced neurotoxicity, albeit rarely life-threatening, often necessitates dose reduction or discontinuation, which may compromise disease control. Although long-term follow-up data are scarce, it has been reported that many breast cancer patients experience persistent numbness of extremities at 12 months after the completion of adjuvant paclitaxel-containing therapy with a frequency of 3–12% (grade 0), 46 –75% (grade 1), 12–21% (grade 2) and 4–8% (grade 3) (Hershman et al, 2011; Tanabe et al, 2013). In addition, a negative impact on the quality of life was documented in affected patients. An imposing challenge in the management of peripheral neuropathy arises from the considerable variation in the time-point of occurrence and the severity of symptoms among treated patients.
Multiple factors influencing peripheral neuropathy development and severity have been reported. The effect of paclitaxel dosing and schedule is well recognised. Generally, paclitaxel-induced peripheral neuropathy occurs more frequently and at greater severity with prolonged treatment duration, higher cumulative dose and higher dose per cycle (Kudlowitz and Muggia, 2013; Miltenburg and Boogerd, 2014). The concomitant use of neurotoxic agents, that is, cisplatin, contributes to the likelihood of development. The addition of either capecitabine or bevacizumab to a taxane, that is, paclitaxel or docetaxel, did not lead to increased rates of taxane-specific toxicities, albeit other toxicities associated with capecitabine (hand-foot syndrome, gastrointestinal toxicities) (O'Shaughnessy and Blum (2006)) or bevacizumab (hypertension, proteinuria and thromboembolic events) (Chan et al, 2010) occurred more frequently. Besides treatment-related factors, patient characteristics predisposing to paclitaxel-induced peripheral neuropathy have been identified and include women, older age, body surface area (BSA) and hyperglycaemia (Kudlowitz and Muggia, 2013; Miltenburg and Boogerd, 2014; Schneider et al, 2015), although their clinical relevance needs further investigation.
Heterogeneity among individuals with respect to susceptibility to paclitaxel-induced peripheral neuropathy may also be attributed to individual genetic differences, among which single-nucleotide polymorphism (SNPs) are the most common form. SNPs in genes involved in paclitaxel metabolic pathways, such as ABCB1, CYP2C8 and CYP3A4, have been shown to modulate paclitaxel pharmacokinetics and may, therefore, be useful for toxicity prediction (Frederiks et al, 2015). CYP2C8*3 comprising c.-416G>A (rs11572080) and c.1196A>G (rs10509681) in perfect linkage disequilibrium has been related to decreased metabolism of paclitaxel and associated with a higher risk of peripheral neuropathy (Gréen et al, 2009; Leskelä et al, 2011; Bergmann et al, 2011a; Hertz et al, 2013). This observation, however, was not consistently reported (Marsh et al, 2007; Bergmann et al, 2011b, 2012; De Graan et al, 2013). Of interest, a newly reported genetic variant of CYP3A4, c.522-191C>T (CYP3A4*22, rs35599367), has been associated with lower enzymatic activity as illustrated by reduced dose requirements of CYP3A4-metabolised drugs, such as statins (Wang et al, 2011). In the context of paclitaxel, De Graan et al (2013) have described that the presence of a CYP3A4 c.522-191 T-allele conferred a higher risk of severe peripheral neuropathy, but the carrier status was not associated with altered pharmacokinetics.
In recent studies, several SNPs possibly involved in paclitaxel-induced neurotoxicity have been identified in genes that play a role in neuronal processes. Leandro-García et al (2012) have found that the highly linked c.-101T>C (rs909964) and c.-112A>G (rs909965) variants in TUBB2A, a microtubule-associated protein, were associated with a lower risk of peripheral neuropathy. Genome-wide association studies (GWAS) have pointed towards SNPs located in genes encoding exchange factors involved in the myelination of peripheral nerves (FGD4) (Baldwin et al, 2012) or proteins related to neuronal development and regeneration (EPHA5) (Baldwin et al, 2012; Leandro-García et al, 2013). Baldwin et al (2012) have identified that FGD4 c.2044-236G>A (rs10771973) was associated with sensory peripheral neuropathy among paclitaxel-treated patients. In two independent GWAS cohorts treated with paclitaxel, EPHA5 c.2895G>A (rs7349683) was found as a predictive marker for peripheral neuropathy (Baldwin et al, 2012; Leandro-García et al, 2013). The functional role of these SNPs in the context of paclitaxel-induced neurotoxicity needs to be characterised.
Further confirmation is necessary to establish the predictive ability of particular SNPs for neurotoxicity among patients receiving paclitaxel. In the present study we analysed a series of candidate SNPs for an association with this adverse event (AE) in a large, well-characterised cohort of HER2-negative metastatic breast cancer (MBC) patients that received first-line paclitaxel-containing chemotherapy.
Materials and methods
Patient cohort
The study population included 188 women with HER2-negative MBC, who were previously enroled in the multicenter, randomised, phase II ATX trial (BOOG2006-06; EudraCT number 2006-006058-83) (Lam et al, 2014). Pharmacogenetic analysis, a preplanned part of the original trial design, was approved by the institutional review board of all participating hospitals and was conducted in accordance with the Declaration of Helsinki. Separate informed consent from patients was required before participation in this study. Blood was collected and serum was stored at −80 °C until DNA isolation.
Treatment
Eligible patients received first-line treatment randomly allocated to the AT arm consisting of paclitaxel (90 mg m−2 on days 1, 8 and 15) and bevacizumab (10 mg kg−1 on days 1 and 8) repeated in a 4-week schedule for 24 weeks, followed by bevacizumab (15 mgkg−1 on day 1) every 3 weeks thereafter, or to the ATX arm consisting of paclitaxel (90 mg m−2 on days 1 and 8), bevacizumab (15 mg kg−1 on day 1) and capecitabine (825 mg m−2 orally twice daily on days 1–14) repeated in a 3-week schedule for 24 weeks, followed by bevacizumab and capecitabine at the same dose every 3 weeks thereafter. Treatment was continued until disease progression, unmanageable toxicity or withdrawal of consent, whichever came first.
Evaluation of peripheral neuropathy and paclitaxel dose modification
Peripheral neuropathy was graded each cycle according to the National Cancer Institute Common Toxicity Criteria for Adverse Events (NCI-CTCAE version 3.0). Dose modification of paclitaxel was based on relevant treatment-related AEs and was documented in case report forms. Paclitaxel dose was reduced to 60 mg m−2 for a second occurrence of grade 2 treatment-related AE or any occurrence of grade 3 treatment-related AEs. Paclitaxel was terminated at third occurrence of grade 2, second occurrence of grade 3 or at first occurrence of a grade 4 AE deemed paclitaxel-related.
Genetic polymorphisms and genotyping
Five polymorphisms in five genes were examined (Table 1). Germline DNA was isolated from 1 ml of serum on a MagNaPure Compact Instrument with a MagNaPure Compact Total Nucleic Acid Isolation kit (Roche Diagnostics, Almere, The Netherlands). Owing to a limited amount of extracted serum DNA, a pre-amplification step was performed with a PreAmp Master Mix Kit according to the manufacturer's instruction (Life Technologies, Bleiswijk, The Netherlands). Genotyping of SNPs CYP2C8*3 (c.416G>A, C__25625794_10, rs11572080), CYP3A4*22 (c.522-191C>T, C_59013445_10, rs35599367), FGD4 (c.2044-236G>A, AH890NA [custom assay], rs10771973) and EPHA5 (c.2895G>A, C___1336545_30, rs7349683) was performed by real-time PCR run on the LightCycler 480II system (Roche Diagnostics) using commercially available predesigned TaqMan probes (Life Technologies). TUBB2A (c.-101T>C, rs909964) genotyping was performed using a Kompetitive Allele Specific PCR (KASP) genotyping assay (LGC Genomics, Teddington, UK). All genotyping was performed in a blinded fashion including water as a negative control. As a quality control, 5% of all DNA samples were measured in duplicate for each genotyping assay. Raw data were analysed with LightCycler 480 software package (release 1.5.0 SP4, Roche Diagnostics).
Table 1. Allele frequency of investigated SNPs.
| Genetic variant | Reference SNP cluster ID | No. of patients | Homozygous wild-type, n (%) | Heterozygous variant, n (%) | Homozygous variant, n (%) | Frequency variant allele in this population | Frequency variant allele (Caucasian)a | Frequency variant allele (Global)a | Call rate, % | HWE P value |
|---|---|---|---|---|---|---|---|---|---|---|
| CYP2C8*3 (c.416G>A) | rs11572080 | 180 | non-carriers/GG 146 (81.1) | *3 carriers/GA 34 (18.9) | AA 0 | 0.094 (A) | 0.11 | 0.065 | 96 | 0.16 |
| CYP3A4 c.522-191C>T | rs35599367 | 172 | CC 154 (89.5) | CT 17 (9.9) | TT 1 (0.6) | 0.055 (T) | 0.05b | 0.022 | 91 | 0.49 |
| TUBB2A c.-101T>C | rs909964 | 182 | TT 100 (54.9) | TC 68 (37.4) | CC 14 (7.7) | 0.26 (C) | 0.26b | 0.43b | 97 | 0.61 |
| FGD4 c.2044-236G>A | rs10771973 | 186 | GG 86 (46.2) | GA 79 (42.5) | AA 21 (11.3) | 0.33 (A) | 0.28 | 0.23 | 99 | 0.66 |
| EPHA5 c.2895G>A | rs7349683 | 172 | GG 77 (44.8) | GA 69 (40.1) | AA 26 (15.1) | 0.35 (A) | 0.40 | 0.31 | 91 | 0.11 |
Abbreviations: HWE=Hardy–Weinberg equilibrium; SNP=Single-nucleotide polymorphism.
Frequency as reported in the SNP Database.
Frequency as reported in 1000 genomes.
Statistical analysis
Data are presented as median with range, actual number and percentage unless otherwise specified. Mann–Whitney U test was used to analyse continuous variables between two groups and the Pearson's χ2 was used for categorical variables. Deviation from the Hardy–Weinberg equilibrium was tested using the Pearson's χ2 test. The relative dose intensity was calculated as the ratio between paclitaxel dose delivered in a time period (mg m−2 week−1) and the planned dose considering no dose modification.
SNPs were assessed univariately by the Kaplan–Meier method and Gehan–Breslow–Wilcoxon test for two toxicity endpoints: (1) cumulative paclitaxel dose (mg m−2) until grade ⩾1 peripheral neuropathy and (2) cumulative paclitaxel dose (mg m−2) until first dose reduction (or study therapy withdrawal) from toxicity deemed unacceptable by treating doctors. Patients not experiencing peripheral neuropathy or dose reduction were censored for the total cumulative dose. SNPs with a P⩽0.10 were selected for multivariate Cox proportional hazards model to estimate hazard ratio (HR) and 95% CI.
The following demographic and clinicopathological factors were examined for the association with the two toxicity endpoints using the Kaplan–Meier method and univariate Cox proportional hazards model: Eastern Cooperative Oncology Group performance status (0 vs 1), age (<65 vs ⩾65 years), BSA (continuous), oestrogen and/or progesterone receptor status (positive vs negative), bone-only disease (yes vs no), previous (neo)adjuvant chemotherapy (yes vs no), previous primary radiotherapy (yes vs no), previous palliative radiotherapy (yes vs no) and the number of metastases (1 vs 2 vs ⩾3). Clinical variables with a P⩽0.10 were considered as covariates in the multivariate analysis. The risk of paclitaxel-related peripheral neuropathy is known to depend on the cumulative dose. Without dose modification, AT-treated patients received a higher cumulative paclitaxel dose than ATX-treated patients (maximum of 1620 vs 1440 mg m−2, respectively). In practice, the total cumulative dose varied widely and there was considerable overlap of patients who received similar cumulative doses regardless treatment arm. To account for between-arm and within-arm differences, patients were categorised into tertile groups of increasing total cumulative dose of paclitaxel and this covariate was included in the multivariate Cox regression analysis.
SNPs were analysed according to the best fitting genetic model in univariate (additive, dominant or recessive) and multivariate analyses (multiplicative). No correction for multiple comparisons was done due to the exploratory nature of this study. All statistical analyses were performed using SPSS version 20 (Chicago, IL, USA). Statistical significance was defined as P⩽0.05.
Results
Study population and paclitaxel treatment
Clinical characteristics of the 188 patients are shown in Table 2. A total of 95 patients were assigned to the AT arm of weekly paclitaxel for 3 weeks on/one week off and 93 patients to the ATX arm of weekly paclitaxel for 2 weeks on/one week off. Demographic and disease characteristics at baseline were well balanced between the treatment arms (Supplementary Table 1). All patients received at least one dose of paclitaxel with a median cumulative dose of 1409 mg m−2 (Table 3). Median relative dose intensity of paclitaxel was 0.94. Owing to the different schedules, as anticipated, the cumulative dose of paclitaxel was higher in AT as compared with ATX (P<0.001). The intended dose of paclitaxel delivered, however, did not differ between the two treatment arms according to the similarity in median relative dose intensities (P=0.28).
Table 2. Baseline patient and demographic characteristics (n=188).
| Characteristic | Study population, n (%) |
|---|---|
| Age, years, median (range) | 57 (32–76) |
| Weight, kg, median (range) | 71 (43–150) |
| Body surface area, m2, median (range) | 1.79 (1.36–2.70) |
| Baseline ECOG performance statusa | |
| 0 | 99 (53) |
| 1 | 89 (47) |
| Oestrogen receptor and/or progesterone receptor | |
| Positive | 164 (87) |
| Negative | 24 (13) |
| Metastases at initial diagnosis | |
| Yes | 25 (13) |
| No | 163 (87) |
| Sites of metastatic disease | |
| Lung metastasis | 63 (34) |
| Liver metastasis | 116 (62) |
| Bone-only disease | 10 (5) |
| No. of metastatic sites | |
| 1 | 23 (12) |
| 2 | 69 (37) |
| ⩾3 | 96 (51) |
| Prior (neo)adjuvant chemotherapy | |
| Anthracycline-containing | 94 (87) |
| CMF | 12 (11) |
| Other | 2 (2) |
| Radiotherapy | |
| Primary breast cancer | 122 (65) |
| Recurrent or metastatic breast cancer | 64 (34) |
| Preexisting diabetes mellitus | |
| Yes | 3 (2) |
| No | 185 (98) |
| Pre-treatment biochemical values of liver and kidney function, median (range) | |
| Aspartate aminotransferase, U l−1 (n=188) | 33 (13–250) |
| Alanine aminotransferase, U l−1 (n=187) | 28 (6–340) |
| Alkaline phosphatase, U l−1 (n=186) | 113 (40–690) |
| Total bilirubin, μmol l−1 (n=186) | 7 (2–27) |
| Albumin, g l−1 (n=181) | 42 (25–51) |
| Blood urea nitrogen, mmol l−1 (n=158) | 4.9 (2.2–15.5) |
| Serum creatinine, μmol l−1 (n=188) | 67 (35–145) |
Abbreviations: CMF=cyclophosphamide, methotrexate and fluorouracil; ECOG=Eastern Cooperative Oncology Group.
Patients with ECOG PS ⩾2 were excluded from clinical trial.
Table 3. Paclitaxel cumulative dose, dose reduction and peripheral neuropathy.
| All patients (n=188) | AT arm (n=95) | ATX arm (n=93) | P valuea | |
|---|---|---|---|---|
| Median cumulative dose, mg m−2 (range) | 1409 (90–1709) | 1519 (120–1709) | 1351 (90–1534) | <0.001 |
| Median relative dose intensityb (range) | 0.94 (0.06–1.18) | 0.94 (0.07–1.18) | 0.94 (0.06–1.18) | 0.28 |
|
Peripheral neuropathy,
n
(%) | ||||
| Grade 1 | 68 (36.2) | 34 (35.8) | 34 (36.6) | |
| Grade 2 | 41 (21.8) | 18 (18.9) | 23 (24.7) | |
| Grade 3 | 17 (9.0) | 9 (9.5) | 8 (8.6) | |
| No peripheral neuropathy | 62 (33.0) | 34 (35.8) | 28 (30.1) | |
| Dose reduction, n (%) | 87 (46) | 42 (44) | 45 (48) | 0.57 |
P value from comparison between AT and ATX arm.
Relative dose intensity was calculated as the ratio between actual paclitaxel dose given in a time period (mg m−2 week−1) and planned dose during 24 weeks of paclitaxel-containing therapy considering no dose modification. Cases of progression or death within 24 weeks were included until the events. Patients taken off study due to toxicity were included until the end of planned paclitaxel-containing therapy.
Paclitaxel-induced peripheral neuropathy and dose reduction
The rates of highest grade peripheral neuropathy experienced per patient are shown in Table 3. There were no cases of grade 4 peripheral neuropathy. A total of 126 patients (67%) experienced grade ⩾1 peripheral neuropathy following the initiation of paclitaxel-based therapy: 64.2% in the AT vs 69.9% in the ATX arm, P=0.41. The rate of dose reduction in the study population was 46% and was similar in both treatment arms (P=0.57). AEs that led to dose reduction of paclitaxel are shown in Supplementary Table 2. There was no significant difference in the rate of grade ⩾1 peripheral neuropathy between patients requiring paclitaxel dose reduction and those without dose reduction (45% vs 47%, P=0.83).
Results of the univariate Cox analysis for clinical variables in relation with two toxicity endpoints are summarised in Supplementary Table 3. Of the demographic and clinicopathological factors, age ⩾65 years was associated with an increased risk of cumulative paclitaxel dose-dependent grade ⩾1 peripheral neuropathy (HR=1.87, 95% CI 1.17–2.97, P=0.008) and was included in the Cox proportional hazards models. BSA has previously been identified as a risk factor for the development of taxane-induced neuropathy (Schneider et al, 2015) and was, therefore, included as a covariate in the analysis of peripheral neuropathy. None of the clinical variables were associated with dose reduction.
Allele frequencies and analyses
The allele frequency of SNPs is shown in Table 1. Although patients' ethnicity was not collected, the vast majority of patients were of Caucasian origin. The allele frequency of SNPs was similar to that reported in the Caucasian population according to the NCBI SNP database (dbSNP) (Sherry et al, 2001). The overall success rate of SNP genotyping was >90% (Table 1). CYP3A4 c.522-191C>T was analysed under a dominant genetic model (CC vs TT+TC), as only one patient was homozygous for the T-allele. The distribution of SNPs in the two treatment arms is presented in Supplementary Table 4.
Genetic polymorphisms and peripheral neuropathy or paclitaxel dose reduction
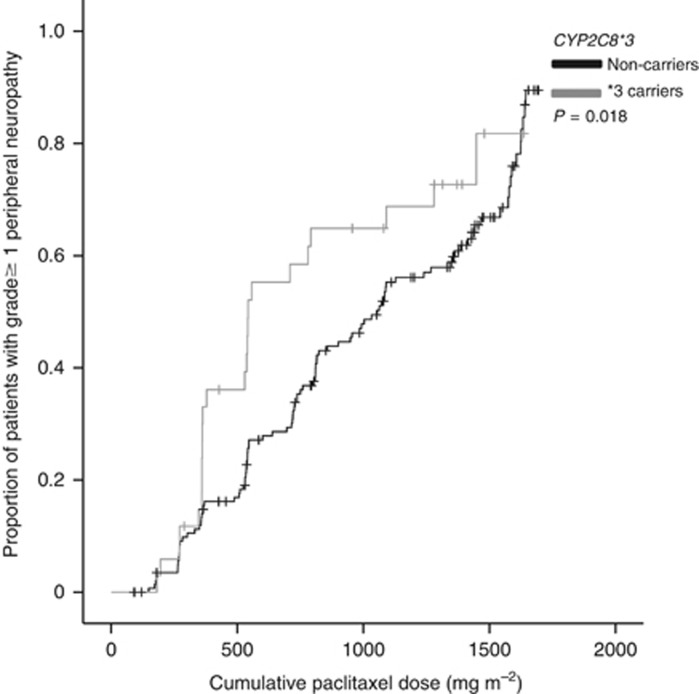
Significant associations in the univariate and multivariate analyses are presented in Table 4. CYP2C8*3 was univariately associated with cumulative dose-dependent peripheral neuropathy. Carriers of a CYP2C8*3 variant received a significantly lower cumulative paclitaxel dose until first onset of grade ⩾1 peripheral neuropathy than non-carriers (Gehan–Breslow–Wilcoxon P=0.018, Figure 1). In the multivariate analysis adjusted for age, BSA and total cumulative paclitaxel dose, CYP2C8*3 variant was independently associated with an increased risk of grade ⩾1 peripheral neuropathy (HR=1.59, 95% CI 1.01–2.52, P=0.045). No significant associations were detected between other SNPs and this peripheral neuropathy endpoint.
Table 4. Analyses of SNPs and possible association with peripheral neuropathy or paclitaxel dose reduction.
|
Peripheral neuropathy |
Paclitaxel dose reduction |
||||||
|---|---|---|---|---|---|---|---|
| Univariate analysis |
Multivariate analysesa,b |
Univariate analysis |
Multivariate analysisa,c |
||||
| Genetic variant | Comparison | P value | Hazard ratio (95% CI) | P value | P value | Hazard ratio (95% CI) | P value |
| CYP2C8*3 (c.416G>A) | *3 carriers vs non-carriers | 0.018 | 1.59 (1.01–2.52) | 0.045 | 0.81 | ||
| CYP3A4 c.522-191C>T | Dominant (TT+CT vs CC) | 0.44 | 0.14 | ||||
| TUBB2A c.-101T>C | Additive | 0.83 | 0.16 | 0.76 (0.53–1.10) | 0.14 | ||
| Dominant (CC+TC vs TT) | 0.66 | 0.10 | |||||
| Recessive (CC vs TC+TT) | 0.59 | 0.13 | |||||
| FGD4 c.2044-236G>A | Additive | 0.62 | 0.10 | 1.38 (1.02–1.86) | 0.036 | ||
| Dominant (AA+AG vs GG) | 0.46 | 0.04 | |||||
| Recessive (AA vs AG+GG) | 0.39 | 0.18 | |||||
| EPHA5 c.2895G>A | Additive | 0.77 | 0.28 | ||||
| Dominant (AA+AG vs GG) | 0.84 | 0.13 | |||||
| Recessive (AA vs AG+GG) | 0.48 | 0.27 | |||||
P value was calculated from the Gehan–Breslow–Wilcoxon test. SNPs with P<0.10 were selected for the Cox proportional hazards analysis.
Multiplicative genetic model, hazard ratio per variant allele except for CYP2C8*3.
Hazard ratio adjusted for age, body surface area, cumulative paclitaxel dose in tertiles.
Hazard ratio adjusted for cumulative paclitaxel dose in tertiles.
Figure 1.
Kaplan–Meier curve of cumulative paclitaxel dose until the occurrence of grade ⩾1 peripheral neuropathy according to CYP2C8*3 (n=180).
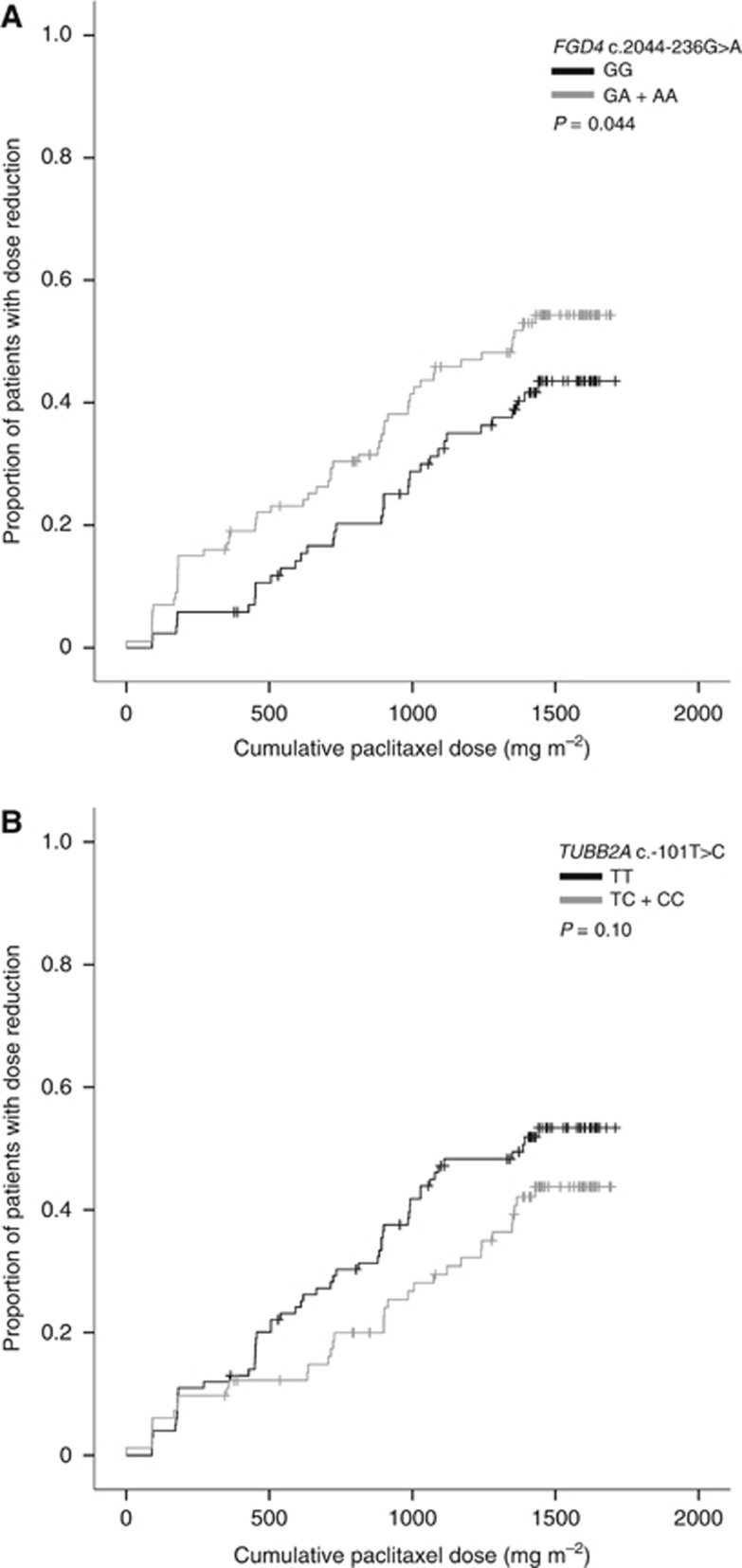
Analysis of the cumulative dose until first paclitaxel dose reduction showed an association for FGD4 c.2044-236G>A. Under a dominant model, the presence of an A-allele was significantly associated with a lower cumulative paclitaxel dose until first reduction compared with the GG genotype (Gehan–Breslow–Wilcoxon P=0.044, Figure 2A). This was associated with an increased risk of paclitaxel dose reduction in the multivariate analysis adjusted for total cumulative paclitaxel dose (HR per A-allele=1.38, 95% CI 1.02–1.86, P=0.036). Interestingly, under a dominant model, the presence of a C-allele in TUBB2A c.-101T>C had a possible association with dose reduction (Gehan–Breslow–Wilcoxon P=0.10, Figure 2B) and was, therefore, selected for calculating the HR. This association with paclitaxel dose reduction was not significant in the multivariate analysis (HR per C-allele=0.76, 95% CI 0.53–1.10, P=0.14).
Figure 2.
SNPs associated with paclitaxel dose reduction. (A) Kaplan–Meier curve of cumulative paclitaxel dose until first dose reduction according to FGD4 c.2044-236G>A (n=186). (B) Kaplan–Meier curve of cumulative paclitaxel dose until first dose reduction according to TUBB2A c.-101T>C (n=182).
Discussion
Pharmacogenetic studies have revealed many candidate SNPs for the prediction of paclitaxel-induced AEs, but reported findings were conflicting. An explanation for this heterogeneity may reside in differences in study design, sample size, definition of toxicity, treatment duration, dose and concomitant neurotoxic medication (Frederiks et al, 2015). In the current study, we focused on a relatively homogeneous group of women with MBC, who were enroled in the phase II ATX trial according to predefined in- and exclusion criteria. All women received a defined dose and a fixed duration of paclitaxel-based therapy. Detailed information with regard to toxicities, dose reduction and received paclitaxel dose were recorded. Therefore, our results may likely provide credible information regarding the predictive value of genetic polymorphisms.
Maximum severity of peripheral neuropathy defined by NCI-CTCAE was used as main study endpoint in most pharmacogenetic studies. Peripheral neuropathy is, however, known to be a cumulative dose-dependent toxicity and occurs earlier in patients receiving higher doses given once every 3 weeks than in lower doses given more frequently (Kudlowitz and Muggia, 2013). We choose grade ⩾1 as a cutoff of this AE, because of its gradual development in the weekly low dose paclitaxel schedule. The cumulative dose until development of toxicity has been employed in the current study and more recent analyses (Leskelä et al, 2011; Baldwin et al, 2012; Leandro-García et al, 2012, 2013; Hertz et al, 2013; Abraham et al, 2014). This approach takes into account the variable treatment duration among advanced cancer patients by censoring those who terminated treatment early due to disease progression or death.
We considered the cumulative dose until first dose reduction of paclitaxel due to a treatment-related AE another clinically meaningful endpoint that generally represents deterioration of the patient's general condition. As paclitaxel treatment is associated with a broad spectrum of clinically relevant toxicities (Supplementary Table 2), it could be speculated that this endpoint captures the overall toxic effects of paclitaxel. Moreover, dose reduction is common practice in oncology allowing symptom alleviation and restoring quality of life. Clinical studies have suggested that dose reduction in chemotherapy may compromise tumour control and survival prospects in treated patients (Lyman, 2009; Loibl et al, 2011), although evidence concerning this topic is scarce. Future evaluation of this study endpoint, in particular for the relationship with clinical outcome, should help to clarity its relevance for pharmacogenetic studies.
The current analysis demonstrated that carriers of a CYP2C8*3 variant (c.416G>A) received a significantly lower cumulative dose of paclitaxel until the development of grade ⩾1 peripheral neuropathy (P=0.018). Multivariate analysis adjusted for age, BSA and differences in the total cumulative paclitaxel dose confirmed an increased risk for CYP2C8*3 variant carriers compared with the non-carriers indicating CYP2C8*3 as a potential marker of paclitaxel-related peripheral neuropathy. CYP2C8 represents a major metabolising enzyme that catalyses the formation of 6α-hydroxypaclitaxel, rendering paclitaxel inactive (Taniguchi et al, 2005). Of interest, CYP2C8*3 (c.416G>A) has been documented to contribute to the attenuation of CYP2C8 activity leading to a decreased clearance of paclitaxel (Bahadur et al, 2002; Gréen et al, 2009). Therefore, although pharmacokinetics was not analysed in the present study, one may speculate that CYP2C8*3 variant carriers are at an increased risk of neurotoxicity as a consequence of increased paclitaxel exposure.
CYP2C8*3 (c.416G>A and/or c.1196A>G) has been examined in previous pharmacogenetic studies on paclitaxel-induced neurotoxicity. Hertz et al (2013) have analysed CYP2C8*3 (c.1196A>G) in a European-American population of 209 women with breast cancer, who received different schedules of (neo)adjuvant paclitaxel-containing therapy. The presence of a CYP2C8*3 allele (c.1196 G) was associated with an increased risk of grade ⩾2 neurotoxicity: each allele had an HR of 1.95 (P=0.031) in a final model that included diabetes history. Subsequently, this association was validated in an African-American cohort (n=107) resulting in an HR of 3.30 (P=0.043) per allele. In a total number of 411 patients consisting of mixed races, the association between CYP2C8*3 (c.1196A>G) and risk of grade ⩾2 neuropathy was still significant (HR 1.98, P=0.004). In their preceding study on 109 patients that were part of the mixed-race cohort, the same group had reported that carriers of a CYP2C8*3 variant (defined as a c.1196 G-allele and/or c.416 A-allele) showed a trend towards a higher risk of grade ⩾3 peripheral neuropathy (P=0.075) (Hertz et al, 2012). In 118 patients with solid tumours receiving a variety of paclitaxel-containing combination schedules, Leskelä et al (2011) have described that CYP2C8*3 (c.416G>A) was significantly associated with the cumulative dose until grade 2 neurotoxicity. In that study, an incremental HR of 1.72 (P=0.032) per variant allele was calculated after adjustment for treatment schedule and age. In another study including 343 patients with mostly breast cancer and ovarian cancer, the CYP2C8*3 genotype (c.416G>A and/or c.1196A>G) was not indicative of maximum severity of peripheral neuropathy, whereas a trend was observed for total paclitaxel dose (mg) until grade >2 peripheral neuropathy in the assessable subset of 146 patients (Lee et al, 2015). Altogether, these and our findings indicate that CYP2C8*3 is important in the occurrence of paclitaxel-induced neurotoxicity.
CYP3A4 represents another major metabolising enzyme of paclitaxel. A novel genetic variant CYP3A4 c.522-191C>T has recently been identified as possible risk factor for paclitaxel-induced neurotoxicity. In the exploratory analysis of 135 males and 126 females treated with different paclitaxel-based schedules, female carriers of a T-allele were at increased risk of developing a higher grade of neuropathy (P=0.043) (De Graan et al, 2013). Validation in 129 males and 110 females also showed that more females than males developed neurotoxicity (P<0.0001). For T-allele carriers an increased risk of development of grade 3 neuropathy was found in both genders, although only 2.5% of patients experienced severe symptoms. We could not confirm CYP3A4 c.522-191C>T as a possible risk factor for neurotoxicity grade ⩾1 in female carriers.
The biological relevance of FGD4, a protein involved in neuronal myelination, is best exemplified by Charcot–Marie–Tooth (CMT) disease, a congenital disease characterised by early onset of motor and sensory neuropathy (Pareyson and Marchesi, 2009). A mutational change in FGD4, among other proteins, is believed to be closely related to the development of CMT. In this regard, genetic variants in FGD4 may be of clinical significance in paclitaxel-induced neurotoxicity. Our analysis indicated that the presence of an A-allele in FGD4 c.2044-236G>A was associated with a decreased paclitaxel cumulative dose until first reduction due to toxicity and an increased risk of dose reduction (HR of 1.38 per A-allele). This finding has not yet been documented in paclitaxel-treated patients. In a recent GWAS of 855 Caucasian patients with primary breast cancer receiving adjuvant dose-dense paclitaxel 175 mg m−2 every 2 weeks × 4 or × 6, FGD4 c.2044-236G>A was associated with a lower cumulative dose until initial onset of grade ⩾2 sensory neuropathy (HR per A-allele=1.57, P=2.6 × 10−6, Baldwin et al, 2012). Although the association did not reach statistical significance at a genome-wide level in the discovery phase, the predictive value of this A-allele was confirmed in two separate cohorts consisting of 154 European (HR per allele=1.72, P=0.013) and 117 African-American (HR per allele=1.93, P=6.7 × 10−3) patients. In our study on weekly paclitaxel 90 mg m−2, the presence of variants in FGD4 c.2044-236G>A was not predictive for grade ⩾1 peripheral neuropathy. The discrepancy with the results of Baldwin et al (2012) might be explained by the different dosing regimens. It has to be mentioned, however, that the association of FGD4 c.2044-236G>A with first dose reduction includes severe cases of peripheral neuropathy among other paclitaxel-related AEs. As no other data exist, the clinical relevance of FGD4 c.2044-236G>A in paclitaxel-induced neurotoxicity deserves further investigation.
Leandro-García et al (2012) have been the first to identify three variants, that is, c.-101T>C, c.-112A>G, and c.-157A>G (rs9501929) located in the TUBB2A promoter that correlated with increased mRNA levels. The c.-101T>C and c.-112A>G variants, in total linkage disequilibrium, conferred an increased transcription rate, whereas the transcriptional activity of the independent c.-157A>G variant was not different from that of the wild-type promoter. The group hypothesised that their observation might provide a biological basis explaining differences in paclitaxel-induced AEs among patients. In the subsequent study in 214 paclitaxel-treated patients, they demonstrated that the presence of the c.-101C/c.-112G variant was protective against developing grade 2 neurotoxicity (HR=0.62, P=0.021) in a multivariate analysis in which a weekly or a 3-weekly schedule was taken into account. There was no association between the c.-157A>G variant and paclitaxel-induced neurotoxicity (Leandro-García et al, 2012). In our study, TUBB2A c.-101T>C was not associated with paclitaxel-induced peripheral neuropathy, but showed an association with paclitaxel dose reduction at a borderline significant level (HR per C-allele=0.70). In contrast to the findings of Leandro-García et al (2012), in a large cohort of 1303 patients with early breast cancer on paclitaxel-based therapy, TUBB2A c.-157A>G was identified with respect to sensory neuropathy (Abraham et al, 2014). The minor allele appeared to be associated with an increased risk of grade ⩾2 sensory neuropathy (odds ratio 1.80, 95% CI 1.20–2.72), whereas the cumulative dose until this endpoint was also lower in carriers (HR 1.60, P=0.005).
Here, we attempted to validate EPHA5 c.2895G>A previously identified in two GWAS as a potentially useful toxicity marker, but no significant association was found in the current study. Ephrin Type-A receptor genes have been incorporated in other analyses. In a cohort of 144 patients with solid tumours treated with paclitaxel and carboplatin, several genetic variants were associated with cumulative dose-dependent grade 2 peripheral neuropathy, although the predefined significance level was not met (Leandro-García et al, 2013). Among them were EPHA4 rs17348202 (HR=4.85, P=1.02 × 10−6) and several genetic variants located in Ephrin Type-A receptors EPHA5, EPHA6 and EPHA8. Baldwin et al (2012) have performed GWAS in 855 breast cancer patients treated with paclitaxel to analyse two toxicity endpoints including the cumulative dose until first onset of grade ⩾2 sensory neuropathy and the maximum severity. For the first endpoint, EPHA5 c.2898G>A showed a borderline significant association (HR=1.63, P<9.6 × 10−7), but this association could not be validated in two independent cohorts. A meta-analysis of data from both GWAS in a total number of 998 patients demonstrated EPHA5 c.2898G>A as a significant risk factor for developing sensory peripheral neuropathy (HR=1.68, P=1.4 × 10−9) (Leandro-García et al, 2013).
Several limitations in the present study warrant discussion. First, patient's ethnicity data were not collected, but most patients were presumed to be of Caucasian origin. This is also likely according to the variant allele frequency resembling that of the population of European descent, that is, CEU population of HapMap (Table 1). Second, we did not account for multiple comparisons given the exploratory or confirmatory nature of this study, because of which results should be interpreted within this context. Third, SNPs with low minor allele frequency may require a large sample size to achieve adequate power for statistical testing. Fourth, patients were treated with weekly paclitaxel 90 mg m−2 given as 3 weeks on/one week off (AT) or as 2 weeks on/one week off (ATX), whereas we combined both patients' groups. The rates of peripheral neuropathy or dose reduction, however, were not different between the two paclitaxel schedules. The multivariate analysis was used to control for the potential confounding effect of differences in the total cumulative dose of paclitaxel, age and BSA. Finally, the call rate of SNP was deemed acceptable (>90%) for genotyping using serum DNA and applying a pre-amplification step. A negative control and technical replicates were included to minimise genotyping error and to ensure accuracy of the results.
In conclusion, several genetic variants have previously been identified to be potentially useful for the prediction of paclitaxel-induced peripheral neuropathy. Results from earlier studies, however, are not always consistent, which may be due to variations in methodologies (Frederiks et al, 2015). In our analysis, we could confirm that CYP2C8*3 was associated with an increased risk of peripheral neuropathy. There was an increased risk of early paclitaxel dose reduction for carriers of the FGD4 c.2044-236 A-allele. These data are of value to add to the current knowledge of factors that might contribute to the differential toxicity from paclitaxel experienced among patients. In particular, our findings point towards FGD4 c.2044-236G>A as a SNP to be incorporated in future pharmacogenetic studies to further assess its role in taxane-related peripheral neuropathy.
Acknowledgments
We thank all patients that participated in this trial and the Dutch Breast Cancer Research Group (BOOG) for trial coordination and data collection. Funding for this work was received from F Hofmann-La Roche Ltd by means of an unrestricted grant.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Presented at the 49th Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, May 31–June 4, 2013.
Supplementary Material
References
- Abraham JE, Guo Q, Dorling L, Tyrer J, Ingle S, Hardy R, Vallier AL, Hiller L, Burns R, Jones L, Bowden SJ, Dunn JA, Poole CJ, Caldas C, Pharoah PPD, Earl HM (2014) Replication of genetic polymorphisms reported to be associated with taxane-related sensory neuropathy in patients with early breast cancer treated with paclitaxel. Clin Cancer Res 20(9): 2466–2475. [DOI] [PubMed] [Google Scholar]
- Bahadur N, Leathart JB, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, Van Houdt J, Hendrickx J, Mannens G, Bohets H, Williams FM, Armstrong M, Crespi CL, Daly AK (2002) CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol 64(11): 1579–1589. [DOI] [PubMed] [Google Scholar]
- Baldwin RM, Owzar K, Zembutsu H, Chhibber A, Kubo M, Jiang C, Watson D, Eclov RJ, Mefford J, McLeod HL, Friedman PN, Hudis CA, Winer EP, Jorgenson EM, Witte JS, Shulman LN, Nakamura Y, Ratain MJ, Kroetz DL (2012) A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALGB 40101. Clin Cancer Res 18(18): 5099–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann TK, Brasch-Andersen C, Gréen H, Mirza M, Pedersen RS, Nielsen F, Skougaard K, Wihl J, Keldsen N, Damkier P, Friberg LE, Peterson C, Vach W, Karlsson MO, Brosen K (2011. a) Impact of CYP2C8*3 on paclitaxel clearance: a population pharmacokinetic and pharmacogenomic study in 93 patients with ovarian cancer. Pharmacogenomics J 11(2): 113–120. [DOI] [PubMed] [Google Scholar]
- Bergmann TK, Brasch-Andersen C, Gréen H, Mirza MR, Skougaard K, Wihl J, Keldsen N, Damkier P, Peterson C, Vach W, Brosen K (2012) Impact of ABCB1 variants on neutrophil depression: a pharmacogenomic study of paclitaxel in 92 women with ovarian cancer. Basic Clin Pharmacol Toxicol 110(2): 199–204. [DOI] [PubMed] [Google Scholar]
- Bergmann TK, Gréen H, Brasch-Andersen C, Mirza MR, Herrstedt J, Hølund B, Du Bois A, Damkier P, Vach W, Brosen K, Peterson C (2011. b) Retrospective study of the impact of pharmacogenetic variants on paclitaxel toxicity and survival in patients with ovarian cancer. Eur J Clin Pharmacol 67(7): 693–700. [DOI] [PubMed] [Google Scholar]
- Chan A, Miles DW, Pivot X (2010) Bevacizumab in combination with taxanes for the first-line treatment of metastatic breast cancer. Ann Oncol 21(12): 2305–2315. [DOI] [PubMed] [Google Scholar]
- De Graan AJ, Elens L, Sprowl JA, Sparreboom A, Friberg LE, Van der Holt B, De Raaf PJ, De Bruijn P, Engels FK, Eskens FA, Wiemer EA, Verweij J, Mathijssen RH, Van Schaik RH (2013) CYP3A4*22 genotype and systemic exposure affect paclitaxel-induced neurotoxicity. Clin Cancer Res 19(12): 3316–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiks CN, Lam SW, Guchelaar HJ, Boven E (2015) Genetic polymorphisms and paclitaxel- or docetaxel-induced toxicities: a systematic review. Cancer Treat Rev 41(10): 935–950. [DOI] [PubMed] [Google Scholar]
- Gréen H, Söderkvist P, Rosenberg P, Mirghani RA, Rymark P, Lundqvist EA, Peterson C (2009) Pharmacogenetic studies of paclitaxel in the treatment of ovarian cancer. Basic Clin Pharmacol Toxicol 104(2): 130–137. [DOI] [PubMed] [Google Scholar]
- Hershman DL, Weimer LH, Wang A, Kranwinkel G, Brafman L, Fuentes D, Awad D, Crew KD (2011) Association between patient reported outcomes and quantitative sensory tests for measuring long-term neurotoxicity in breast cancer survivors treated with adjuvant paclitaxel chemotherapy. Breast Cancer Res Treat 125(3): 767–774. [DOI] [PubMed] [Google Scholar]
- Hertz DL, Motsinger-Reif AA, Drobish A, Winham SJ, McLeod HL, Carey LA, Dees EC (2012) CYP2C8*3 predicts benefit/risk profile in breast cancer patients receiving neoadjuvant paclitaxel. Breast Cancer Res Treat 134(1): 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz DL, Roy S, Motsinger-Reif AA, Drobish A, Clark LS, McLeod HL, Carey LA, Dees EC (2013) CYP2C8*3 increases risk of neuropathy in breast cancer patients treated with paclitaxel. Ann Oncol 24(6): 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlowitz D, Muggia F (2013) Defining risks of taxane neuropathy: insights from randomized clinical trials. Clin Cancer Res 19(17): 4570–4577. [DOI] [PubMed] [Google Scholar]
- Lam SW, De Groot SM, Honkoop AH, Jager A, Ten Tije AJ, Bos MM, Linn SC, Van den Bosch J, Kroep JR, Braun JJ, Van Tinteren H, Boven E (2014) Paclitaxel and bevacizumab with or without capecitabine as first-line treatment for HER2-negative locally recurrent or metastatic breast cancer: a multicentre, open-label, randomised phase 2 trial. Eur J Cancer 50(18): 3077–3088. [DOI] [PubMed] [Google Scholar]
- Leandro-García LJ, Inglada-Peréz L, Pita G, Hjerpe E, Leskelä S, Jara C, Mielgo X, González-Neira A, Robledo M, Åvall-Lundqvist E, Gréen H, Rodríguez-Antona C (2013) Genome-wide association study identifies ephrin type A receptors implicated in paclitaxel induced peripheral sensory neuropathy. J Med Genet 50(9): 599–605. [DOI] [PubMed] [Google Scholar]
- Leandro-García LJ, Leskelä S, Jara C, Gréen H, Åvall-Lundqvist E, Wheeler HE, Dolan ME, Inglada-Perez L, Maliszewska A, de Cubas AA, Comino-Méndez I, Mancikova V, Cascon A, Robledo M, Rodríguez-Antona C (2012) Regulatory polymorphisms in beta-tubulin IIa are associated with paclitaxel-induced peripheral neuropathy. Clin Cancer Res 18(16): 4441–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Apellaniz-Ruiz M, Johansson I, Vikingsson S, Bergmann TK, Brosen K, Green H, Rodriguez-Antona C, Ingelman-Sundberg M (2015) Role of cytochrome P450 2C8*3 (CYP2C8*3) in paclitaxel metabolism and paclitaxel-induced neurotoxicity. Pharmacogenomics 16(9): 929–937. [DOI] [PubMed] [Google Scholar]
- Leskelä S, Jara C, Leandro-García LJ, Martínez A, García-Donas J, Hernando S, Hurtado A, Vicario JC, Montero-Conde C, Landa I, López-Jiménez E, Cascon A, Milne RL, Robledo M, Rodríguez-Antona C (2011) Polymorphisms in cytochromes P450 2C8 and 3A5 are associated with paclitaxel neurotoxicity. Pharmacogenomics J 11(2): 121–129. [DOI] [PubMed] [Google Scholar]
- Loibl S, Skacel T, Nekljudova V, Luck HJ, Schwenkglenks M, Brodowicz T, Zielinski C, von MG (2011) Evaluating the impact of relative total dose intensity (RTDI) on patients' short and long-term outcome in taxane- and anthracycline-based chemotherapy of metastatic breast cancer—a pooled analysis. BMC Cancer 11: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman GH (2009) Impact of chemotherapy dose intensity on cancer patient outcomes. J Natl Compr Canc Netw 7(1): 99–108. [DOI] [PubMed] [Google Scholar]
- Marsh S, Paul J, King CR, Gifford G, McLeod HL, Brown R (2007) Pharmacogenetic assessment of toxicity and outcome after platinum plus taxane chemotherapy in ovarian cancer: the Scottish Randomised Trial in Ovarian Cancer. J Clin Oncol 25(29): 4528–4535. [DOI] [PubMed] [Google Scholar]
- Miltenburg NC, Boogerd W (2014) Chemotherapy-induced neuropathy: a comprehensive survey. Cancer Treat Rev 40(7): 872–882. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy JA, Blum JL (2006) Capecitabine/taxane combination therapy: evolving clinical utility in breast cancer. Clin Breast Cancer 7(1): 42–50. [DOI] [PubMed] [Google Scholar]
- Pareyson D, Marchesi C (2009) Diagnosis, natural history, and management of Charcot–Marie–Tooth disease. Lancet Neurol 8(7): 654–667. [DOI] [PubMed] [Google Scholar]
- Schneider BP, Li L, Radovich M, Shen F, Miller KD, Flockhart DA, Jiang G, Vance G, Gardner L, Vatta M, Bai S, Lai D, Koller D, Zhao F, O'Neill A, Smith ML, Railey E, White C, Partridge A, Sparano J, Davidson NE, Foroud T, Sledge GW Jr (2015) Genome-wide association studies for taxane-induced peripheral neuropathy in ECOG-5103 and ECOG-1199. Clin Cancer Res 21(22): 5082–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29(1): 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, Hashimoto K, Shimizu C, Hirakawa A, Harano K, Yunokawa M, Yonemori K, Katsumata N, Tamura K, Ando M, Kinoshita T, Fujiwara Y (2013) Paclitaxel-induced peripheral neuropathy in patients receiving adjuvant chemotherapy for breast cancer. Int J Clin Oncol 18(1): 132–138. [DOI] [PubMed] [Google Scholar]
- Taniguchi R, Kumai T, Matsumoto N, Watanabe M, Kamio K, Suzuki S, Kobayashi S (2005) Utilization of human liver microsomes to explain individual differences in paclitaxel metabolism by CYP2C8 and CYP3A4. J Pharmacol Sci 97(1): 83–90. [DOI] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W (2011) Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 11(4): 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.