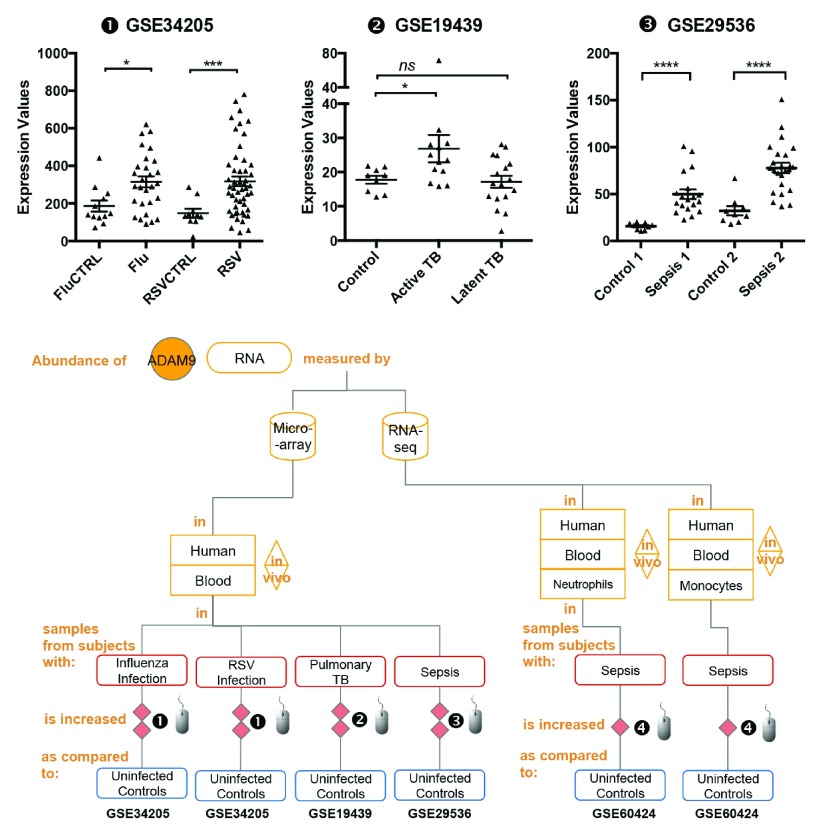
Figure 2. The abundance of ADAM9 increases during infection.
mRNA expression levels for ADAM9 was measured by microarrays in whole blood obtained from children hospitalized with acute RSV and influenza virus infection (GSE34205), pulmonary tuberculosis patients (GSE19439) and patients with sepsis (GSE29536).
The graphical legend represents visually the information associated with the public datasets used for the meta-interpretation of ADAM9 transcriptional profiles. The flow chart indicates how data were generated. Diamonds indicate availability of supporting data and in the interactive version are hyperlinked to context-rich interactive plots. Links to these plots are also provided below:
❶ GSE34205: In this study gene expression profiles were obtained from the whole blood of critically ill pediatric patients 23, Children hospitalized with acute RSV and influenza virus infection were offered study enrollment after microbiologic confirmation of the diagnosis. Blood samples were collected within 42–72 hours of hospitalization. Median age of subjects was 2.4 months (range 1.5–8.6). Uninfected subjects of similar demographics were recruited in the study and served as age-matched controls. Children with suspected or proven polymicrobial infections, with underlying chronic medical conditions (i.e congenital heart disease, renal insufficiency), with immunodeficiency, or those who received systemic steroids or other immunomodulatory therapies were excluded. As stated in the manuscript: “The Institutional Review Boards at the University of Texas Southwestern Medical Center and Baylor Institute for Immunology Research approved this study, and informed consent was obtained from legal guardians prior to any study-related procedure.” More details are available via the interactive data browsing application under the “study” tab.
https://gxb.benaroyaresearch.org/dm3/miniURL/view/Ka
❷ GSE19439: Whole blood was collected from patients with different spectra of tuberculosis (TB) disease and healthy controls 25. All patients were sampled prior to the initiation of any anti-mycobacterial therapy. Active Pulmonary TB: all patients confirmed by isolation of Mycobacterium tuberculosis on culture of sputum or bronchoalvelolar lavage fluid. Latent TB: All patients were positive by tuberculin skin test (>14mm if BCG vaccinated, >5mm if not vaccinated) and were also positive by Interferon-Gamma Release assay (IGRA). As stated in the manuscript: “The local Research Ethics Committees (REC) at St Mary’s Hospital, London, UK approved the study”.
https://gxb.benaroyaresearch.org/dm3/miniURL/view/Kb
❸ GSE29536: Whole blood was collected from culture positive patients meeting criteria for sepsis enrolled in two independent cohorts (Sepsis 1 and Sepsis 2) 22. Uninfected controls recruited in this study were of similar demographics. As stated in the manuscript: “The study was performed by recruitment of patients who were suspected of having hospital or community acquired infection. Clinical specimens were collected for bacterial culture within 24 hours following the diagnosis of sepsis. All blood samples were obtained at the Khon Kaen Regional Hospital, Khon Kaen, Thailand as approved by Khon Kaen University Ethic Committee for Human Research (Project number HE470506)”.
https://gxb.benaroyaresearch.org/dm3/miniURL/view/Jl
❹ GSE60424: Whole blood sample of healthy donors, patients during acute infections (meningococcal sepsis, E. coli sepsis, C. difficile colitis), multiple sclerosis patients pre- and 24 hours post- interferon treatment, patients with Type 1 diabetes and patients with ALS were obtained and monocyte, neutrophil, CD4 T cell, CD8 T cells, B cell, NK Cell isolated prior to profiling via RNA sequencing 21.
https://gxb.benaroyaresearch.org/dm3/miniURL/view/Kc
Statistical significance was determined using Mann-Whitney U test. ns, not significant, * p < 0.05, *** p < 0.001 and *** p < 0.0001. The horizontal lines indicate mean ± standard errors (SE).