Figure 2. Development of rational-design principles.
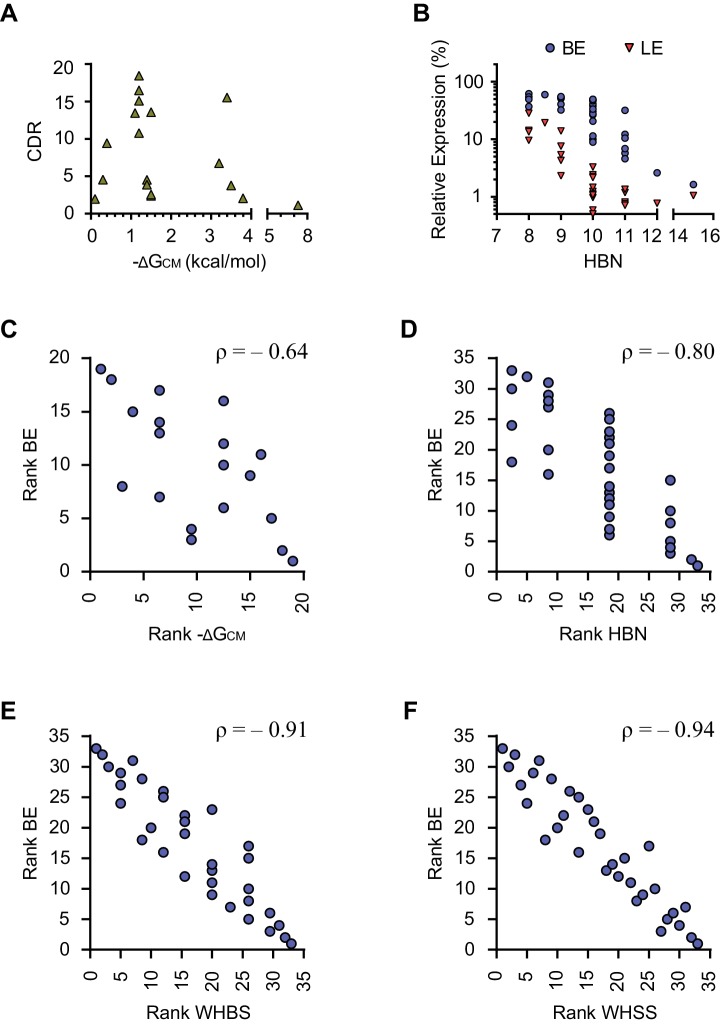
(A) The predicted hybrid free energies of the communication modules (ΔGCM) of test-panel aptazymes were calculated with RNAstructure Web Server (Bellaousov et al., 2013). BE (blue circles) and LE (red triangles) were then plotted against the negative ΔGCM value for all aptazymes whose ΔGCM could be determined. Note the poor correlation between ΔGCM and BE or LE. (B–C) BE and LE of the indicated aptazyme variants are shown. Expression levels are normalized to that of the CNTL control construct. The communication-module sequences of these variants are displayed to the left of the figures. Red indicates base pairs different from the consensus. Note that all constructs in each set have the same base pairs, and that order of these pairs impacts BE. (D) Accordingly, every CM base pair was assigned a weight based on its proximity to the ribozyme, with distal bases assigned lower weights. A Weighted Hydrogen Bond Score (WHBS) was then calculated as the weighted sum of hydrogen bonds in the communication module. BE (blue circles) and LE (red triangles) of each variant were plotted against WHBS. Note that WHBS better correlates with the rank order of BE than does ΔGCM or the unweighted sum of hydrogen bond numbers in the communication module (Figure 2—figure supplement 1B). (E–F) The BE and LE of the indicated aptazymes are shown, with their communication-module sequences indicated to the left. Nucleotides with the potential to form inter-strand base-stacking interactions are highlighted as red. (G) Potential inter-strand purine base-stacking interactions were weighted by proximity to the ribozyme, and added to the WHBS to form a modified score (WHSS). BE (blue circles) and LE (red triangles) were plotted against WHSS of the corresponding aptazyme variant. Note that the WHSS better correlates with the rank order of BE than does the WHBS or ΔGCM. An inflection point of the BE at WHSS 6.7 is indicated with a dashed line. Shading indicates a region with WHSS values 6.7 ± 0.3. (H) The CDR values of all 32 test-panel aptazymes were plotted against their WHSS. Note the CDR optimum near the WHSS value of 6.7 (shaded). Aptazymes that exhibited the highest (Tc12, Tc15) or lower-than-expected (Tc31 and Tc32) CDRs are indicated. (I–K) The CDRs of the indicated aptazymes are shown, with their communication-module sequences displayed to the left. Note that aptazymes in each panel share identical sequences (I) or similar WHSS (J,K), but differ in the stability of the communication-module base pair immediately proximal to the Tc aptamer (red). Aptazyme WHSS values range from 6.13 to 6.29 in (J), and from 6.58 to 6.63 in (K). Tc31 and Tc32, with 1 and 0 hydrogen bonds at this position, are outliers in Figure 2H, likely because these weak bonds destabilize the aptamer, and lower its affinity for Tc. (L) Efficient aptazymes met two criteria: a communication-module WHSS value within the range of 6.7 ± 0.3, and an aptamer-proximal communication-module base pair with stability similar to or higher than that of the original aptamer. Aptazymes that meet both criteria have high BE and CDR ('optimal' aptazymes), whereas those that fail to meet one have low BE or narrow CDR ('suboptimal' aptazymes). The CDR and BE of the both optimal and suboptimal aptazymes are ordered by CDR and plotted. Data points in panels A, D, G, H and L represent mean of three biological replicates. Data points in panels B, C, E, F, I, J, and K represent mean ± S.D. of three biological replicates. Numerical data for all the figures are available in Figure 2—source data 1.
DOI: http://dx.doi.org/10.7554/eLife.18858.005