Fig. 1.
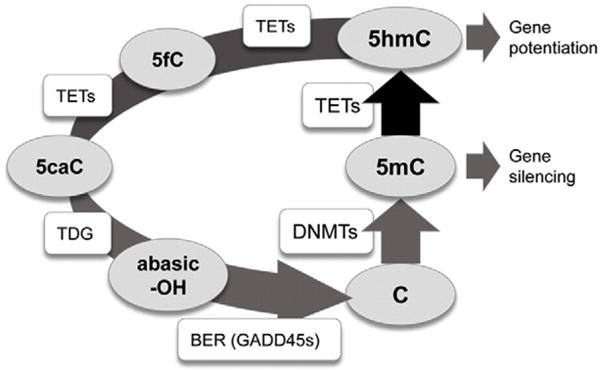
The dynamic modification cycle of DNA cytosine and its impact on gene activity. This model of the turnover of modified cytosine (C) residues emphasizes the central role of DNMTs in the methylation of C to 5-methylcytosine (5mC) and TETs in the rate limiting removal of 5mC by oxidation to 5-hydroxymethylcytosine (5hmC). The dynamic turnover of 5mC appears critical to regulating rapid changes in linked gene expression (Meagher, 2014; Wu and Zhang, 2014). TETs may further oxidize 5hmC to 5-formalcytosine (5fC) and 5-carboxycytosine (5caC). Thymine DNA glycosidase TDG removes the modified 5fC or 5caC bases leaving an abasic nucleotide (–OH). Base excision repair (BER) repairs the single nucleotide gap in double stranded DNA back to a C residue. Enzymes are in square boxes and nucleotide bases are in ovals. The diagram was modified from (Kohli and Zhang, 2013), based on the data in (Chen et al., 2012; Ramon et al., 2012; Dubois-Chevalier et al., 2014; Haseeb et al., 2014; Oger et al., 2014).
