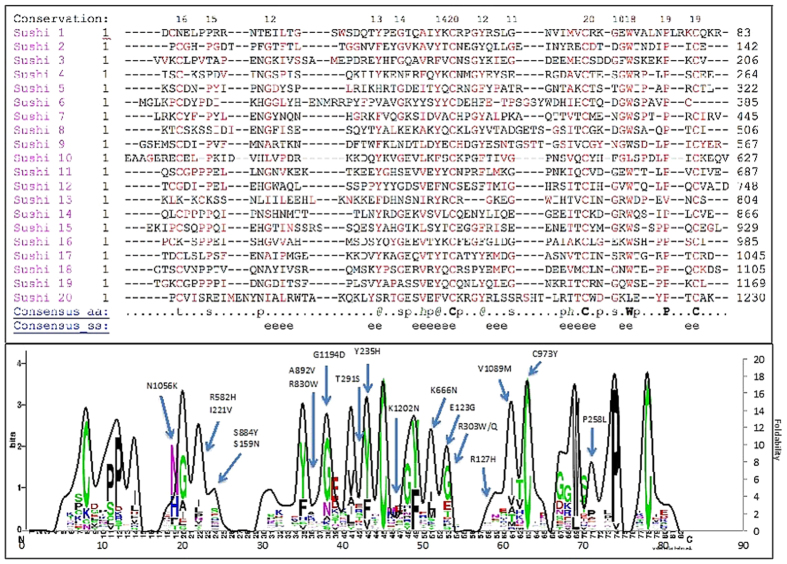
Figure 4. Multiple sequence alignment showing the conservation of the unfolding propensities of the complement factor H sushi domains.
The 20 sushi domains were separated, and the sequences of each domain were compared by Promals3D multiple sequence alignment. (a) The alignment of the sushi domains shows significant conservation for the residues with high foldability values. These residues, which could cause protein unfolding when mutated, are colored red. (b) From the sequence alignment shown in (a), the logos of the conserved residues (http://weblogo.berkeley.edu/logo.cgi) were calculated and superimposed on the average unfolding values, shown by the black curve. The highest peaks of the curve correspond to the positions of the red residues from the sequence alignment in (a). The previously described genetic mutations64 are labeled on the plot.