Fig. 7.
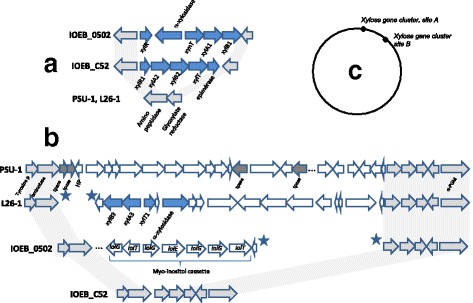
Schematic representation of the 3 xylose gene cassettes of O. oeni. a Xylose cluster in O. oeni IOEB_C52 (GC rate in the cluster 37.2%) and IOEB_0502 (GC: 37.1%) b Xylose cluster in O. oeni IOEB_L26-1 (GC: 37.9%). c Insertion sites of xylose cassettes in O. oeni chromosome. The genes putatively associated with xylose metabolism appear as blue arrows and the dark gray arrows represent genes that may be at the origin of the acquisition of the xylose genes. The stars indicate ends of contigs (draft genomes). XylA: xylulose kinase; XylB xylose isomerase; XylT: permease; XynT xyloside transporteur; XylR: transcription regulator; Tpase: transposase, HP: hypothetical protein, PGM: phosphoglucomutase, aminotase: aminotransferase
