Figure 1.
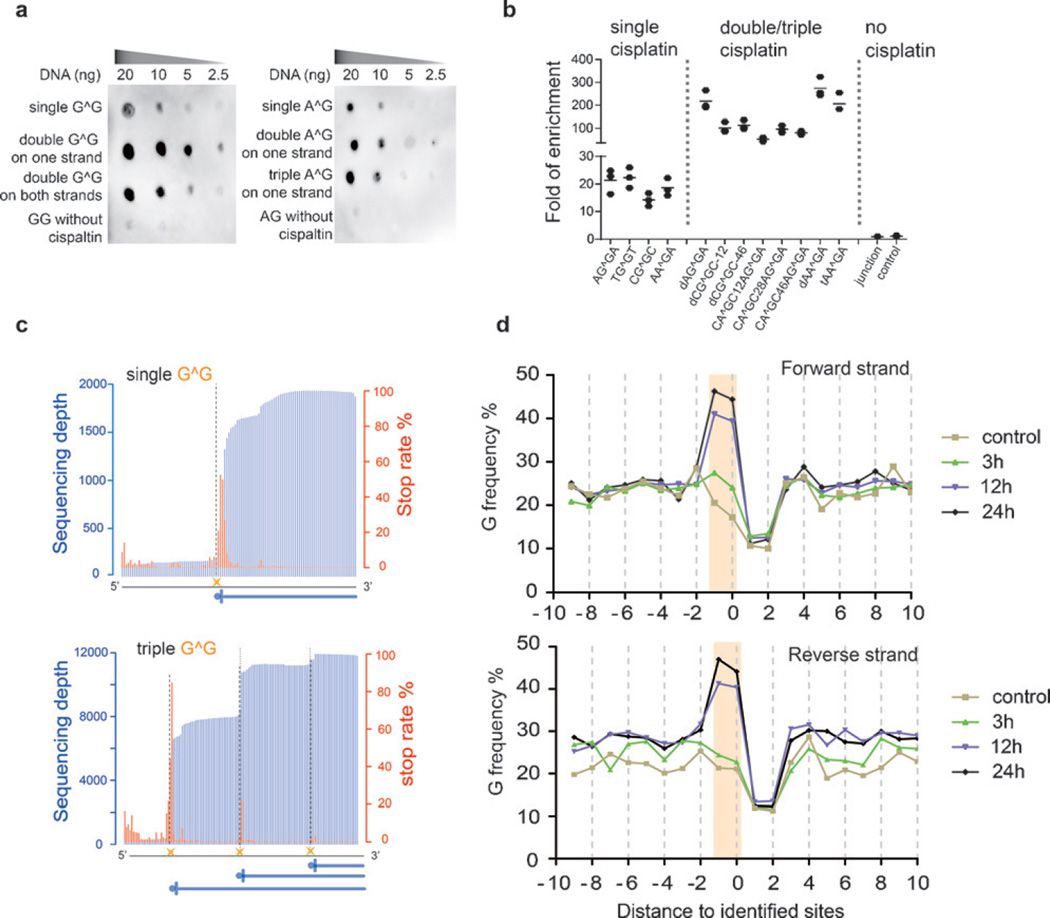
Cisplatin-seq reliably detects cisplatin–DNA adducts in model DNA and the human genome. a) Dot blot analysis showed that HMGB1 domain A specifically binds to cisplatin-modified model DNA sequences (“G^G” denotes cis-GG 1,2-intrastrand adducts; all sequences in this Figure have two adenosines flanking the G^G and A^G adducts). b) Enrichment of cisplatin-modified model DNA sequences after HMGB1 domain A pull-down experiments (n = 3). For the sequence named “dCG^GC-12”, “d” stands for “double” cis-GG adducts, and “12” means a 12 bp distance between two cis-GG adducts. Similarly, in other sequences, “t” stands for “triple” and the number represents the distance between two nearby cisplatin adducts; “junction” represents a sequence with a four-way junction structure, which was used as a control. c) The stop rate information in model sequences was used to detect cisplatin crosslinking sites at base resolution. The blue vertical lines are sequencing depths whereas the red lines are calculated stop rates (see the Supporting Information). The dashed lines represent the cisplatin sites. The sequences in this Figure have two cytidines flanking the G^G adducts. d) Frequency of guanine nucleosides spanning the identified sites for treatment for 3 h, 12 h, and 24 h. “Position 0” corresponds to sites of high stop rates.