Abstract
Objectives:
The objective of this article is to investigate the contribution of colon and blood CD4+ T-cell subsets expressing the chemokine receptor CCR6 to HIV persistence during antiretroviral therapy.
Design:
Matched sigmoid biopsies and blood samples (n = 13) as well as leukapheresis (n = 20) were collected from chronically HIV-infected individuals receiving antiretroviral therapy. Subsets of CD4+ T cells with distinct differentiation/polarization profiles were identified using surface markers as follows: memory (TM, CD45RA−), central memory (TCM; CD45RA−CCR7+), effector (TEM/TM; CD45RA−CCR7−), Th17 (CCR6+CCR4+), Th1Th17 (CCR6+CXCR3+), Th1 (CCR6−CXCR3+), and Th2 (CCR6−CCR4+).
Methods:
We used polychromatic flow cytometry for cell sorting, nested real-time PCR for HIV DNA quantification, ELISA and flow cytometry for HIV p24 quantification. HIV reactivation was induced by TCR triggering in the presence/absence of all-trans retinoic acid.
Results:
Compared with blood, the frequency of CCR6+ TM was higher in the colon. In both colon and blood compartments, CCR6+ TM were significantly enriched in HIV DNA when compared with their CCR6− counterparts (n = 13). In blood, integrated HIV DNA levels were significantly enriched in CCR6+ versus CCR6− TCM of four of five individuals and CCR6+ versus CCR6− TEM of three of five individuals. Among blood TCM, Th17 and Th1Th17 contributed the most to the pool of cells harboring integrated HIV DNA despite their reduced frequency compared with Th2, which were infected the least. HIV reactivation was induced by TCR triggering and/or retinoic acid exposure at higher levels in CCR6+ versus CCR6− TM, TCM, and TEM.
Conclusion:
CCR6 is a marker for colon and blood CD4+ T cells enriched for replication-competent HIV DNA. Novel eradication strategies should target HIV persistence in CCR6+CD4+ T cells from various anatomic sites.
Keywords: antiretroviral therapy, CCR6, central memory CD4+ T cells, HIV reservoirs, retinoic acid, Th17, viral outgrowth assay
Introduction
Antiretroviral therapy (ART) has transformed the HIV-1 epidemic into a manageable chronic disease [1–3]. However, HIV reservoirs persist in resting CD4+ T cells [3–11] and viral rebound occurs upon ART interruption [12,13] despite low frequencies of infected CD4+ T cells [3,14–18]. Given the challenges related to lifelong ART treatment such as adherence, viral escape, toxicity, and cost, combined with suboptimal drug penetration into anatomic sanctuaries [19–21], the identification of rare cell subsets that carry HIV reservoirs based on surface markers and the implementation of latency reversal strategies represent major research priorities [3,14,15,17,22–25].
HIV reservoirs are defined as long-lived latently infected cells that allow persistence of replication-competent HIV for years during ART [3,15,17]. The HIV and simian immunodeficiency virus (SIV) exploit the long-lived properties of central memory (TCM) and transitional memory CD4+ T cells to ensure its persistence [26–29]. Effector memory (TEM) CD4+ T cells also harbor replication-competent HIV in ART-treated individuals [27,30], but their contribution may be lower compared with TCM[31]. Moreover, follicular helper T cells are permissive to infection [32] and represent an important HIV reservoir during ART [33]. Furthermore, the finding that HIV reservoirs also persist in long-lived stem cell memory CD4+ T cells [30,34] raised new challenges for HIV eradication strategies. Apart from their heterogeneity in terms of differentiation stage and anatomic localization, memory CD4+ T cells exhibit distinct polarization profiles tailored to their antigenic specificities [35–37]. In addition to Th1 and Th2, Th17 cells represent a third major lineage of CD4+ T cells specialized in maintaining immunity against pathogens at mucosal surfaces, including the gut [38,39]. HIV/SIV efficiently replicate in Th17 as demonstrated by studies by our group and others [40–48]. More specifically, mucosal Th17 cells represent major targets of HIV/SIV infection [49,50] and subsequent rapid depletion [51–57]. Consistent with this paradigm, a very recent study identified CCR6+RORgt+ Th17 as the first cells to be infected upon vaginal SIV challenge [58]. The developmental plasticity [59,60], combined with the long-lived properties of Th17 cells [61,62], recently raised new questions relative to their potential contribution to HIV persistence during ART. Although the contribution of blood Th17 to HIV reservoir persistence during ART is supported by recent studies by our group [63] and others [64,65], this aspect remains poorly investigated at mucosal sites. Further studies in this area are important to orient new cell subset-targeted eradication strategies.
Herein, we investigated the contribution of colon and blood memory CD4+ T-cell subsets expressing the chemokine receptor CCR6 to HIV persistence during ART and the presence of replication-competent HIV in these cells.
Material and methods
Study participants
HIV-infected individuals with undetectable plasma viral load receiving ART (<40 copies HIV RNA/ml; HIV+ on ART (B20-B32), one elite controller (EC1; CD4 cell counts: 753 cells/μl; time since infection: 258 months; untreated: 61-year old), and uninfected volunteers were recruited at the McGill University Health Centre (Table 1). Sigmoid biopsies (≈32 biopsies/donor) were collected during colonoscopy. Biopsies were processed using Liberase DL (Roche Diagnostics, Mannheim, Germany), as previously described [66,67]. Matched peripheral blood samples (20 ml/donor) were collected from biopsy donors the same day. Finally, leukaphereses were collected from other HIV+ individuals on ART (ART #01–19) and uninfected individuals (Table 1), as previously reported [68]. Plasma viral load was measured using the Amplicor HIV-1 monitor ultrasensitive method (Roche) (Roche Diagnostics Systems, Branchburg, New Jersey, USA). The date of infection was estimated using clinical and laboratory data as well as patient history information. Peripheral blood mononuclear cells were isolated by gradient density centrifugation [41,43] and frozen.
Table 1.
Demographic and clinical characteristics of study cohorts.
| Study cohorts | |||||
| Matched blood/sigmoid biopsies | Leukapheresis | ||||
| HIV+ (B20–32; n = 13) | HIV− (n = 7) | HIV+ (ART01–19; n = 20) | HIV− (n = 13) | ||
| Age (years) | Median | 55 | 60 | 46 | 45 |
| Range | 50–67 | 54–69 | 24–62 | 26–58 | |
| Sex | M/F | 13/0 | 5/2a | 15/5 | 12/1 |
| Race | C/H/AA | 12/1/0 | 7/0/0 | 16/0/4 | 13/0/0 |
| CD4+ cell counts (cells/μl) | Median | 679 | 750 | 575 | 745 (n = 12b) |
| Range | 175–985 | 456–1229 | 269–886 | 375–1424 (n = 12b) | |
| CD8+ cell counts (cells/μl) | Median | 914 | 320 | 470 | 362 (n = 12b) |
| Range | 489–1117 | 158–582 | 240–1272 | 95–709 (n = 12b) | |
| CD4+/CD8+ ratios | Median | 0.67 | 2.11 | 1.15 | 2.38 (n = 12b) |
| Range | 0.16–2.01 | 1.83–2.89 | 0.49–2.00 | 1.56–3.98 (n = 12b) | |
| Time since infection (months) | Median | 233 | N/A | 153 | N/A |
| Range | 48–348 | N/A | 48–288 | N/A | |
| ARTc | Yes | N/A | Yes | N/A | |
| Time of aviremiad on ART (months) | Median | 72 (n = 10b) | N/A | 50 (n = 17b) | N/A |
| Range | 24–165 (n = 10b) | N/A | 11–85 (n = 17b) | N/A | |
AA, African-American; ART, antiretroviral therapy; C, Caucasian; F, female; H, Hispanic; M, male; N/A, not applicable.
aOne F was positive for Hepatitis A and B viruses.
bData available on a fraction of donors only.
cART consisted of a combination of two nucleoside reverse transcriptase inhibitors with either one nonnucleoside reverse transcriptase inhibitors, protease inhibitor, or integrase inhibitor.
dPlasma viral load <40 HIV RNA copies/ml.
Ethics statement
The study using sigmoid biopsies and blood from HIV-infected and uninfected study participants was conducted in compliance with the principles included in the Declaration of Helsinki and received approval from the Institutional Review Board of the McGill University Health Centre and CHUM-Research Centre. All study participants signed a written informed consent for their study participation.
Flow cytometry analysis
The following fluorochrome-conjugated antibodies (Abs) were used for polychromatic flow cytometry analysis: CD3-Pacific-blue (UCHT1), CD4-Alexa-Fluor 700 (RPA-T4), CCR4-PE-Cy7 (1G1), CCR6-PE (11A9), CCR7-PE-Cy7 (3D12), CXCR3-PE-Cy5 (1C6) (BD Biosciences, San Diego, California, USA), CCR7-FITC (150503) (R&D Systems, Minneapolis, Minnesota, USA), CD45RA-APC-eFluor780 (HI100) (eBioscience, San Diego, California, USA), CD3-Alexa700 (UCHT1), CD326-BV650 (9C4), CD8-PerCP-Cy5.5 (RPA-T8), CD19-PerCP-Cy5.5 (HIB19), CD66b-PerCP-Cy5.5 (G10F5), human leukocyte antigen - antigen D related (HLA-DR)-BV785 (L243; Biolegend ,San Diego, California USA), CD4-FITC (SFCI12T4D11), and HIV-p24-RD1 (FH190–1–1); (Beckman Coulter, Fullerton, California, USA). Live/Dead Fixable Aqua Dead Cell Stain Kit (Vivid; Life Technologies, Burlington, Ontario, Canada) was used to exclude dead cells. Cells were analyzed by fluorescence-activated cell sorting (FACS) using the BD-LSRII cytometer and BD-Diva (BD Biosciences) and FlowJo (Tree Star, Inc., Ashland, Oregon, USA) softwares. All Abs were titrated for an optimal noise/signal ratio and Ab cocktails were validated by comparing single with multiple staining. Positive gates were placed based on fluorescence minus one controls [41,69].
Cell sorting
CD4+ T cells were sorted by magnetic-activated cell sorting and FACS, as previously described [41,43,45,47]. Briefly, cells stained with Abs were suspended (40 × 106 cells/ml) in sorting media (PBS 1× with 5% fetal bovine serum (FBS) and 25 mmol/l Hepes buffer) and sorted using the BD-FACSAriaIII (BD Biosciences). The sorting gates were set using fluorescence minus one controls [41,69]. The viability staining Vivid (Life Technologies) was included in each staining cocktail to exclude dead cells. Sorted subsets were in average more than 99% pure based on CD4, CD45RA, and CCR6 expression and more than 90% based on CCR7, CCR4, and CXCR3 expression (Suppl. Figures 2 and 4).
RT-PCR for CCR6 mRNA detection
Total RNA was isolated using RNeasy kit (Qiagen Inc., Hilden, Germany), as previously described [45,47]. One-step reverse transcription PCR (RT-PCR) (Qiagen Inc., Toronto Ontario, Canada) using CCR6 Quantitect primers (Qiagen Inc., Toronto) was carried out in an Eppendorf Mastercycler PCR System (Applied Biosystems, Foster City, California, USA). The products of RT-PCR amplifications were visualized on a 1% agarose gel. Reactions were performed in the presence and absence of reverse transcriptase to distinguish between genomic DNA and mRNA amplification.
HIV DNA quantification
The quantification of total and integrated HIV DNA was performed by nested real-time PCR using specific primers and amplification conditions, as we previously described [27,41,43,45,47].
HIV reactivation assay
Cells were cultured in 48-well plates at 2 × 106 cells/well/ml in the presence of immobilized CD3 and soluble CD28 Abs (1 μg/ml). Cells were cultured in the presence or absence of all-trans retinoic acid (ATRA, 10 nmol/l). At days 3, 6, and 9, media was refreshed with RPMI 1640 containing FBS (10%) and IL-2 (5 ng/ml) and/or ATRA. HIV p24 levels were quantified using a homemade ELISA assays [27,41,43,45,47]. The intracellular expression of HIV p24 was quantified by flow cytometry upon staining with appropriate Abs using the Cytofix/Cytoperm kit (BD Biosciences).
IL-17A ELISA
IL-17A levels in cell culture supernatants were quantified by a specific ELISA assay (eBiosciences).
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 5 software (GraphPad Software, Inc., La Joya, California, USA). Briefly, paired t-test P values were calculated for the comparison between two sets of matched samples, Friedman with Dunn's multiple comparisons tests were applied to determine statistical significance for differences between multiple matched cell subsets, and Mann–Whitney P values were calculated for comparisons between unmatched groups.
Results
Colon and blood memory CCR6+ T cells are enriched in HIV DNA
To study the contribution of CD4+ T-cell subsets expressing the chemokine receptor CCR6 to HIV persistence during ART, matched sigmoid biopsies and blood samples were collected from n = 13 ART-treated chronically HIV-infected individuals with undetectable plasma viral load (<40 HIV RNA copies/ml; Table 1; B20-B32) and EC1. Memory CCR6+ and CCR6− CD3+CD4+CD45RA− T cells negative for CD8, CD19, CD66b, and CD326 were analyzed for frequency and HLA-DR expression and sorted by flow cytometry for HIV DNA quantification (Fig. 1a). The frequency of CD3+, CD3+CD4+, and CD3+CD4+CCR6+ T cells was similar in the colon of HIV± on ART compared with uninfected individuals (Table 1); nevertheless, the frequency of blood CD3±CD4± T cells was significantly reduced (Suppl. Figure 1A-C), consistent with differences in the CD4 cell counts between the two cohorts (Table 1). The frequency of memory CCR6+ T cells was significantly higher in the colon versus blood of both HIV+ on ART (Fig. 1b) and uninfected controls (Suppl. Figure 1C). An increased expression of the activation marker HLA-DR was observed on memory CD3+CD4+ T cells expressing CCR6 versus CCR6− T cells from the colon but not the blood of HIV+ on ART (Fig. 1c and d), with similar differences being observed in uninfected individuals (Suppl. Figure 1D-E). The pool of CCR6+ T cells in the gut compared with blood was enriched in cells with a CXCR3+ phenotype (Fig. 1e), suggesting a Th1Th17 polarization of these cells [41,70]. These findings indicate that the colon is a preferential site for CCR6+ T-cell infiltration and that these cells are activated in the colon regardless of the HIV status.
Fig. 1.
Phenotypic analysis and HIV DNA quantification in colon and blood CCR6+ and CCR6− T-cell subsets.
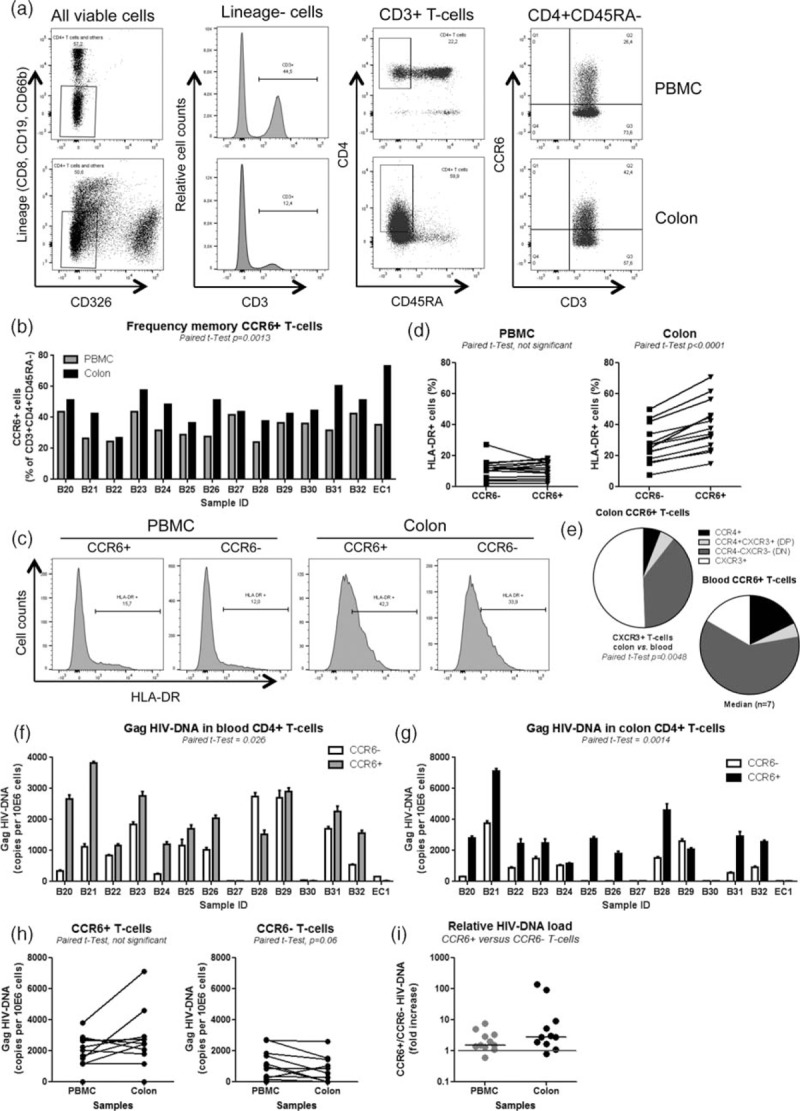
ART, antiretroviral therapy; PBMC, peripheral blood mononuclear cells. Memory (CD45RA−) CD3+CD4+ T-cell subsets positive or negative for CCR6 were isolated from matched colon biopsies and fresh PBMC of n = 13 HIV-infected individuals with undetectable plasma viral load (<40 HIV RNA copies/ml) receiving ART and one elite controller (Table 1; B20-32; elite controller 1). Briefly, cells were stained with a cocktail of Abs against CD3, CD4, CCR6, CCR4, CXCR3, HLA-DR as well as the lineage markers CD326 (epithelial cells), CD8 (CD8+ T cells), CD19 (B cells), and CD66b (granulocytes). Shown is (a) the gating strategy for the flow cytometry analysis/sorting of memory CCR6+ and CCR6− CD3+CD4+CD45RA− T cells; (b) the frequency of CCR6+ subsets in sigmoid biopsies and PBMC; and (c and d) the expression of HLA-DR on CCR6+/CCR6− subsets from sigmoid biopsies versus PBMC in one representative donor (c) and statistical analysis in all donors (d). Shown in (e) is the frequency of cells expressing CCR4 and/or CXCR3 within the pool of CCD6+ T cells in colon versus blood of n = 7 HIV+ individuals on ART. (a–e) (f–i) Sorted, matched colon and PBMC samples were used for Gag HIV DNA quantification. Shown are HIV DNA levels in CCR6+ versus CCR6− cells from PBMC, (f) and colon biopsies (g) (mean ± SD of triplicates), as well as comparisons of HIV DNA levels in CCR6+ and CCR6− T cells between blood and colon (h) and the relative HIV DNA load in CCR6+ versus CCR6− T cells (fold increase) from BPMC and colon (i). Paired t-test values are indicated on the graphs.
Memory CCR6+ and CCR6− T cells from matched sigmoid biopsies and blood were further sorted and levels of total HIV DNA (Gag, late reverse transcripts) were quantified by nested real-time PCR [27,41]. Results in Fig. 1f and g demonstrate significantly higher levels of HIV DNA in CCR6+ versus CCR6− T cells from both blood (P = 0.026; median 1.5-fold increase) and colon (P = 0.0014; median 2.8-fold increase). Although statistical significance was not reached, a fraction of donors exhibited higher HIV DNA levels in CCR6+ T cells from the colon versus blood, whereas HIV DNA levels in CCR6− T cells from the colon versus blood were reduced (Fig. 1h). Of note, HIV DNA levels were low to undetectable in blood and colon CCR6+/CCR6− T cells of the EC1 and two ART-treated individuals (B37, B30; Fig. 1f and g), in part because of limited numbers of sorted cells (typically between 3000 and 50 000 cells/subset/PCR). These results reveal that CCR6+ compared with CCR6− T cells are highly enriched in the colon where they exhibit a superior state of activation and significantly contribute to the persistence of HIV DNA during ART in both colon and blood compartments.
Previous studies documented a transient downregulation of CCR6 expression upon T-cell receptor (TCR) triggering [71], raising the possibility that in HIV-infected study participants with chronic systemic immune activation, the CCR6− fraction may be contaminated by formerly CCR6+ T cells. To test this possibility, highly pure total memory CCR6+/CCR6− T cells sorted by flow cytometry (Suppl. Figure 2A) were analyzed for the expression of CCR6 mRNA by RT-PCR. In all donors, CCR6 mRNA was detected at higher levels in CCR6+ versus CCR6− subsets, with CCR6 mRNA being detectable in the CCR6- fractions of only two of five study participants (Suppl. Figure 2B). Thus, in some HIV-infected donors, CCR6− T cells likely include a small fraction of CCR6+ T cells that downregulated CCR6 probably as a consequence of their activation in vivo.
Central memory CCR6+ T cells are major contributors to integrated HIV DNA reservoirs in the blood
We next studied the frequency and size of the HIV DNA reservoir in blood CCR6+/CCR6− T cells with TCM (CD45RA−CCR7+) and effector/transitional memory (TEM/TM, CD45RA−CCR7−) phenotypes, using well established surface markers [37]. Cell subsets were isolated from large numbers of peripheral blood mononuclear cells obtained by leukapheresis from aviremic HIV-infected study participants receiving ART for more than 11 months (Table 1; ART#01–19). Total CD4+ T cells from a subset of donors were first screened for the presence of total and integrated HIV DNA using nested real-time PCR [27,41]. Levels of total versus integrated HIV DNA levels were always higher but the two measurements were positively correlated (Suppl. Figure 3). Using the gating strategy depicted in Fig. 2a, a reduced frequency of CCR6+ TCM and most significantly CCR6+ TEM/TM was observed in HIV+ on ART compared with HIV− controls (Fig. 2b). Highly pure T-cell subsets were sorted by FACS (Fig. 2c) from n = 5 HIV+ individuals on ART (Table 1) and used for the quantification of integrated HIV DNA. Despite typical donor-to-donor variations in the absolute numbers of integrated HIV DNA copies/subset, the highest levels of integrated HIV DNA were observed in CCR6+ TCM in 4/5 donors (Friedman P = 0.0031, n = 5; Fig. 2d and e). When the integrated HIV DNA copy numbers were normalized relative to the proportion of each cell subset, a tendency for a greater contribution of CCR6+ TCM to the pool of proviral reservoir was observed, compared with other subsets (Fig. 2f).
Fig. 2.
Significant contribution of CCR6+ TCM to the pool of HIV DNA reservoir.
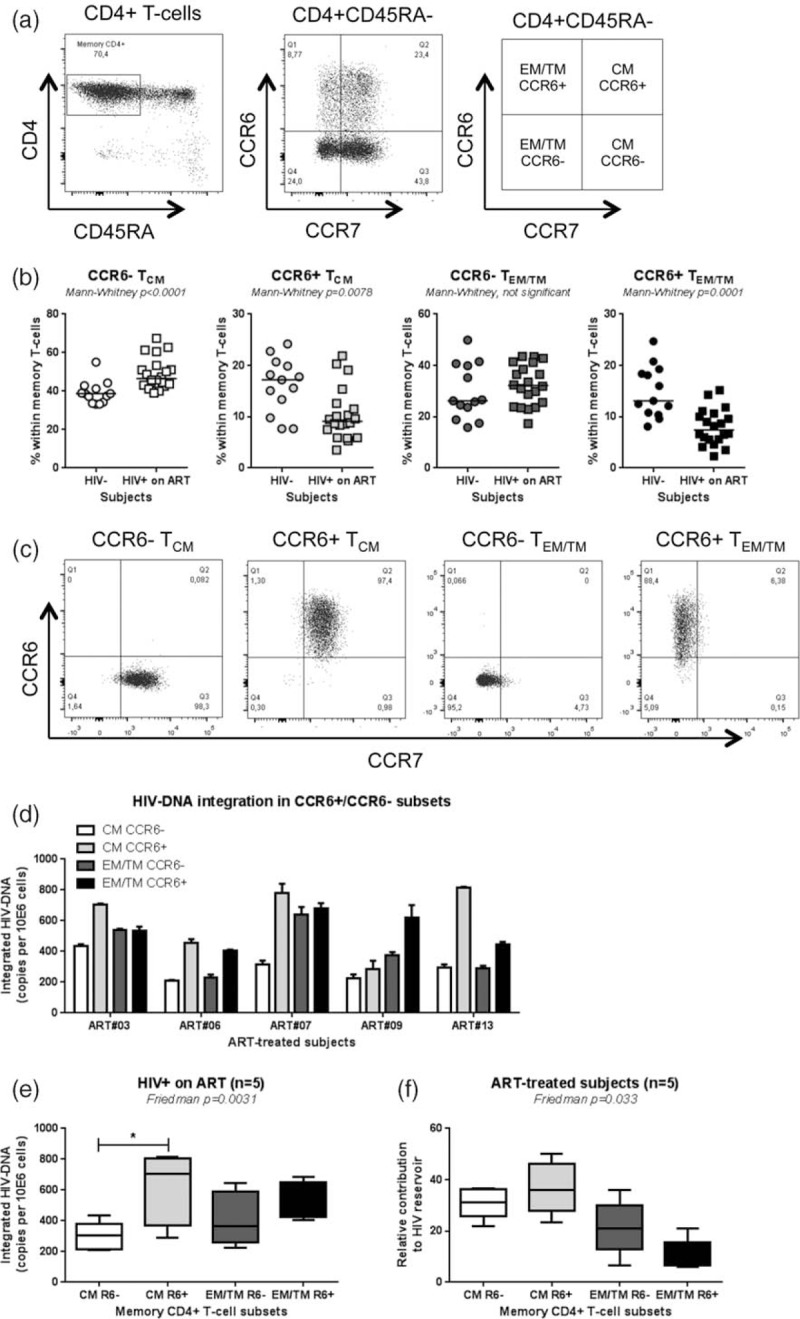
ART, antiretroviral therapy; TCM, central memory T cell; TEM/TM, effector T cell. Peripheral blood mononuclear cell from HIV+ on ART individuals and uninfected controls (HIV−) were stained with a cocktail of CD3, CD4, CD45RA, CCR6, and CCR7 Abs. Shown are (a) the gating strategy for the identification of CCR6+ and CCR6− T cells with TCM (CD45RA−CCR7+) and TEM/TM (CD45RA−CCR7−) cells and (b) the frequency of these subsets in HIV− (n = 13) versus HIV+ on ART (n = 20) individuals. Further, CCR6+ and CCR6− T cells with TCM and TEM/TM phenotypes were sorted by flow cytometry from HIV+ on ART individuals and integrated HIV DNA levels were quantified. Shown are (c) the cell purity after sort and (d and e) levels of integrated HIV DNA in subsets isolated from n = 5 HIV+ on ART individual (mean ± SEM). (f) The relative contribution of CCR6+/CCR6− TCM and TEM/TM to the pool of integrated HIV DNA within total memory T cells was calculated considering the frequency of each subset as following: [relative pool of HIV-infected cells per subset (HIV DNA copies per 10e6 cells for each subset × subset frequency/100)/pool of HIV-infected cells per total memory T cells (sum of the relative pool of HIV-infected cells in all four subsets) × 100]. Indicated in the graphs are Mann–Whitney (b) and Friedman and Dunn's multiple comparison test P values (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001) (e and f).
To further identify discrete T-cell subsets that contribute to HIV DNA persistence during ART, CCR4 and CXCR3 were used to identify TCM with a Th17 (CCR6+CCR4+CXCR3−) and Th1Th17 (CCR6+CCR4−CXCR3+) polarization profile, as well as Th1 (CCR6−CCR4−CXCR3−) and Th2 (CCR6−CCR4+CXCR3−) [41,70] (Fig. 3a). The frequency of TCM with Th2 and Th17 but not Th1 and Th1Th17 phenotypes was significantly diminished in HIV+ on ART compared with HIV− individuals (Fig. 3b; Table 1). The four TCM subsets were sorted by flow cytometry from n = 5 HIV+ individuals on ART, as previously described [41] (Suppl. Figure 4) and levels of integrated HIV DNA were quantified. Among TCM cells, Th17 and Th1Th17 subsets harbored significantly higher levels of integrated HIV DNA compared with Th2 cells (Fig. 3c and d). Further, when the contribution to the pool of HIV reservoir was calculated relative to the proportion of cells per subset, TCM with Th1Th17 as well as Th1 phenotypes contributed the most to the HIV DNA pool, whereas Th2 TCM contributed the least (Fig. 3e). Although, differences in the relative contribution of Th17 versus Th2 to HIV reservoir did not reach statistical significance using the Dunn's multiple comparisons test, likely as a consequence of donor-to-donor variation in absolute HIV DNA levels as well as Th17 frequency (Fig. 3e), matched comparisons between Th17 and Th2 showed statistically significant differences (paired t-test P = 0.0036).
Fig. 3.
Significant contribution of TCM with Th17 and Th1Th17 phenotypes to the pool of HIV DNA reservoir.
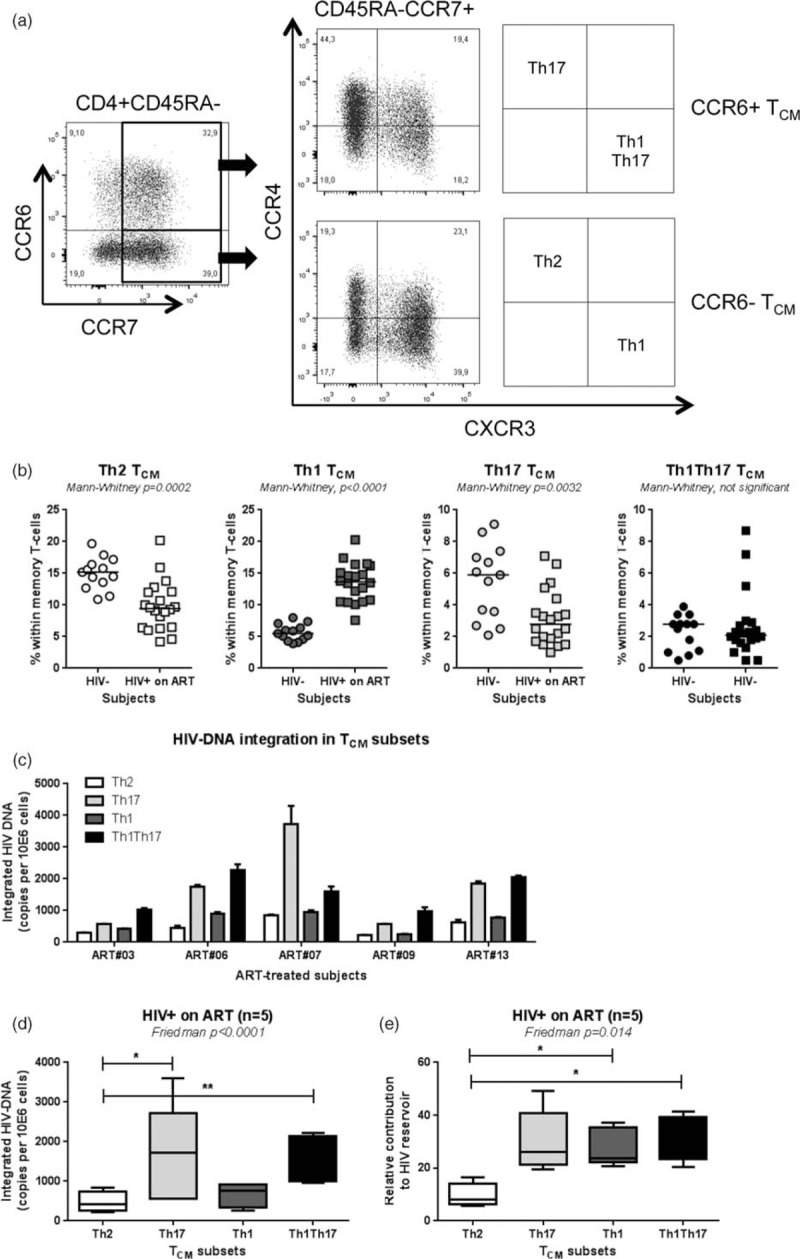
ART, antiretroviral therapy; TCM, central memory T cell. Peripheral blood mononuclear cell from HIV+ on ART and HIV individuals were stained with a cocktail of CD3, CD4, CD45RA, CCR6, CCR4, CCR7, and CXCR3 Abs. (a) Shown is the gating strategy for the identification of TCM cells with Th17 (CCR6+CCR4+CXCR3−), Th1Th17 (CCR6+CCR4−CXCR3+), Th2 (CCR6−CCR4+CXCR3−), and Th1 (CCR6−CCR4−CXCR3+) phenotypes. The frequency of the four TCM subsets was compared between HIV+ on ART and HIV− controls (b). Highly pure TCM subsets were sorted by flow cytometry from HIV+ on ART individuals (Suppl. Figure 3) and integrated HIV DNA levels were quantified. Shown are levels of integrated HIV DNA in subsets from n = 5 individual donors (c) and statistical analysis in all donors (d). The relative contribution of the four TCM subsets to the pool of integrated HIV DNA within TCM cells was calculated considering the frequency of each subset as described in Fig. 2f legend. (e). Indicated in the graphs are Mann-Whitney (b) and Friedman and Dunn's multiple comparison test P values (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001) (d and e).
Together these results reveal the important although not exclusive contribution of CCR6+ TCM cells with Th17 and Th1Th17 polarization phenotypes to the persistence of integrated HIV DNA during ART, despite their decreased frequency in the peripheral blood of HIV+ individuals on ART.
HIV reactivation occurs in subsets of memory CD4+ T cells expressing CCR6
We finally addressed the question whether CCR6+ T-cell subsets are enriched in replication-competent HIV. TCR triggering leads to optimal HIV reactivation in CD4+ T cells [24,72]. Also, we previously demonstrated that ATRA increases HIV permissiveness in CCR6+ T cells in vitro[43]. To determine whether ATRA directly regulates the activity of the HIV promoter, pilot experiments were performed with HeLa Human cervical carcinoma cells (TZM-BL) cells, engineered to carry the luciferase gene under the control of HIV promoter, as well as in ACH2 cells [a human T cell line derived from a leukemia donor (A3.01) infected with HIV] harboring one copy of integrated HIV DNA per cell. Increased HIV promoter activity was observed in the presence of ATRA when TZM-BL cells were infected with replication-competent HIV or transfected with HIV-Tat (Suppl. Figure 5A-B) and HIV p24 levels were significantly increased in phorbol 12-myristate 13-acetate-treated ACH2 cells (Suppl. Figure 5C). Therefore, for an optimal HIV reactivation, T cells were stimulated with CD3/CD28 Abs and cultured in the presence or absence of ATRA, in the absence of ART, with IL-2 added at day 3 postculture (Fig. 4a). In contrast to the standard viral outgrowth assays (VOAs) [14], no target cells were added. Viral replication was measured by HIV p24 quantification by ELISA and flow cytometry. The Th17-specific effector cytokine IL-17A was almost exclusively detected in cell culture supernatants of the CCR6+ TM, TCM, and TEM/TM fractions (Fig. 4b), indicative that contamination by activated T cells that downregulated CCR6 expression was minor. Consistent with their preferential infection (Figs. 1–3), HIV reactivation occurred preferentially in CCR6+ versus CCR6− TM, TCM, and TEM/TM subsets in 3/3 study participants in the presence or absence of ATRA, as determined by the HIV p24 levels measured by ELISA in culture supernatants (Fig. 4c and d) and FACS quantification of HIV p24+ cell frequency (Fig. 4e and f). Of note, the effect of ATRA was more robust on CCR6+ TEM/TM compared with TM and TCM subsets, and HIV reactivation failed in CCR6+ TCM of ART #15, whereas in the same donor HIV reactivation could be detected in TM and TEM/TM subsets (Fig. 4c–f). Together, these results provide evidence that the pool of memory CD4+ T cells carrying replication-competent HIV DNA is highly heterogeneous, that CCR6 is a marker for cells preferentially infected, and that ATRA may be used together with TCR triggering to outgrow HIV more efficiently in ART-treated study participants.
Fig. 4.
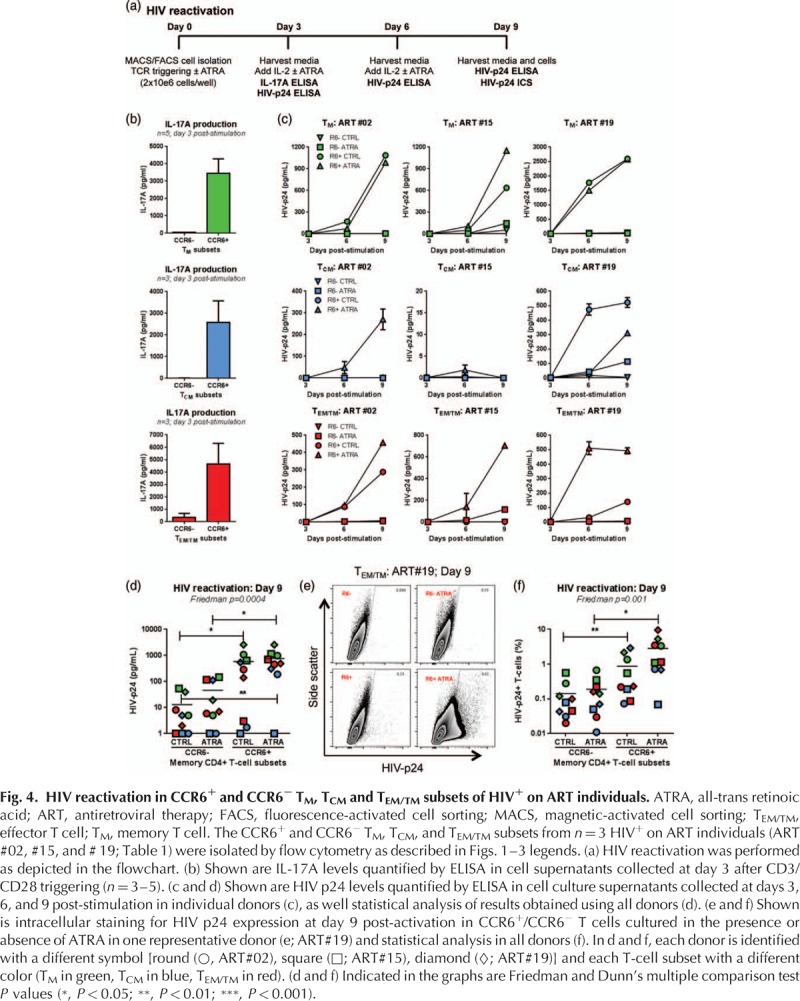
Discussion
In this study, we demonstrate that memory CD4+ T-cell subsets expressing the chemokine receptor CCR6 are enriched in HIV DNA in both colon and blood of HIV-infected individuals receiving ART. We also demonstrated that blood CCR6+ T cells with TCM and Th17 and/or Th1Th17 phenotypes were enriched in integrated HIV DNA; and that HIV reactivation is induced more robustly in CCR6+ versus CCR6− TM, TCM, and TEM, upon TCR triggering in the presence of ATRA. These findings are consistent with the concept that fractions of Th17 cells are long lived [61,62,63] and support HIV reservoir persistence during ART [63,64,65]. HIV uses the molecular machinery of the host cells for integration into specific sites [73]. Whether the integration landscape of HIV differs in CD4+ T-cell subsets with unique transcriptional profiles, such as CCR6+ T cells, and whether this leads to distinct mechanisms of HIV latency and reactivation remains to be determined in future studies.
CCR6 regulates cell migration into various anatomic sites including the intestinal mucosa [74–76]. CCR6 expression on CD4+ T cells is associated with the Th17 lineage commitment [41,70,77]. Although not all CCR6+ T cells produce IL-17, a major fraction of CCR6+IL-17A− T cells become IL-17A+ upon exposure to specific signals in vitro[78]. This is consistent with the most recent Th17 polarization model that includes two distinct steps, specification and acquisition of effector functions [79]. Studies by our group and others previously demonstrated preferential HIV replication in memory CD4+CCR6+ T cells producing IL-17A [40–48]. The superior HIV permissiveness of CCR6+ versus CCR6− T cells is explained by the relatively high expression of the HIV coreceptor CCR5 and the expression of multiple postentry HIV permissiveness factors [41–45,47]. Consistently, CCR6+RORγt+ Th17 cells were identified very recently as the first targets of SIV infection upon vaginal challenge [58]. HIV infection per se, in addition to other possible mechanisms (e.g., altered trafficking into mucosal sites [75,80], paucity of Th17 precursors [81,82] and Th17-polarizing dendritic cell subsets [83], IL-21 deficit [84]), contribute to Th17 depletion from gut-associated lymphoid tissues thus causing dramatic alterations of intestinal barrier function [51–57]. ART fails to restore Th17 frequency/function at mucosal sites unless treatment is initiated during the very first days/weeks of infection [85–87]. Indeed, early ART initiation is the only strategy to reduce the size of HIV reservoirs [18,26]. Our study provides original evidence that CCR6+ T cells are enriched in frequency in the colon versus blood of HIV+ on ART individuals and carry higher levels of HIV DNA compared with their CCR6− counterparts. Our results also revealed that colon versus blood CCR6+ T cells exhibit an activated (HLA-DR+) phenotype. This raises the possibility that residual viral replication may occur in colon CCR6+ T cells during ART; however, the size of our sorted cell samples was too small to explore this possibility. HLA-DR expression on colon CCR6+ T cells was observed in both infected/uninfected individuals but whether this is the consequence of the interaction with local microbiota [88], remains to be investigated. Of note, colon compared with blood CCR6+ T cells were enriched in cells expressing CXCR3, corresponding to a Th1Th17 phenotype [70] that is associated with the highest levels of CCR5 expression [41,42]. Finally, access to large leukapheresis samples allowed us to further explore the validity of CCR6 as a marker for cells enriched in HIV DNA. Despite the reduced frequency of cells expressing CCR6 in the blood of HIV+ on ART compared with uninfected individuals, we observed that total CCR6+ TCM population, as well as CCR6+CCR4+ (Th17 phenotype) and/or CCR6+CXCR3+ (Th1Th17 phenotype) TCM, contributed most significantly to the pool of integrated HIV DNA. In contrast, cells with a Th2 phenotype (CCR4+CCR6−CXCR3−) carried the lowest levels of HIV DNA, likely because of their lack of CCR5 expression [41].
Our findings are consistent with those recently published by Sun et al.'s [64] and Khoury et al.'s [65] groups for peripheral blood CCR6+ T cells. All these findings are in line with a model in which the CCR6–CCL20 axis plays a critical role HIV latency establishment in resting CD4+ T cells [89] and support the need for early intervention to block HIV infection of CCR6+ T cells. Such strategies may include microbicides such as glycerol monolaurate, a compound that proved to be efficient in stopping SIV dissemination from the portal sites of entry in a simian model of infection [90]. Other strategies may be inspired from the most current understanding of molecular mechanisms regulating HIV permissiveness in CCR6+ Th17 [45,47,48,91], to prevent new infection and HIV persistence in these cells during ART.
The ‘gold standard’ method for the quantification of replication-competent HIV reservoirs is the VOA [14,24]. Given the limited number of memory CD4+CCR6+ T cells, we could isolate using leukapheresis samples of HIV+ on ART individuals, we established a modified VOA in which cells were stimulated via CD3/CD28 and cultured in the presence or absence of ATRA. We previously demonstrated that ATRA increases HIV replication specifically in CCR6+ T cells via entry (CCR5 upregulation) and postentry mechanisms [43]. Of note, retinoic acid (RA)-responsive elements (RARE) are present within the HIV long terminal repeat [92], supporting a direct effect of ATRA on the HIV promoter. This possibility is consistent with very recent studies that used acitretin, a second-generation retinoid, to kill latently infected cells upon HIV reservoir reactivation [93]. In addition to a potential therapeutic application [93], we propose the use of ATRA in VOA to detect replication-competent HIV in small biological samples isolated from patients upon various therapeutic interventions. By using this VOA, we demonstrated that HIV productive replication occurs more efficiently in memory CCR6+ compared with CCR6− T cells.
One potential bias in our study is linked to the possibility that regulatory T cells (Tregs) are present in the CCR6+ T-cell fractions. Indeed, Tregs were reported to express CCR6, CCR4, and/or CXCR3 [82,94], thus insuring their colocalization with Th17 subsets at various anatomic sites. In humans, Tregs can be identified as CD3+CD4+ T cells with a CD25highCD127lowFoxP3+ phenotype [81,95]. We previously reported that the expression of FoxP3 mRNA was minimal in CCR6+ T-cell subsets isolated from leukapheresis [41]. Nevertheless, considering the increased frequency of Tregs to the detriment of Th17 cells at mucosal sites [75,96], the ability of Tregs to support HIV replication [97], as well as the ability to Th17 cells to transdifferentiate into Tregs [60], future studies are needed to distinguish the contribution of CCR6+ Th17 versus CCR6+ Tregs to HIV reservoir persistence during ART. Such studies will require larger biological samples from organ ectomy or autopsy tissues; there is also the need of using reliable Treg markers and functional assays for the identification of Tregs with suppressive activity, as FoxP3 expression was documented to be upregulated in conventional T cells upon TCR triggering [98].
One discrepancy in our study is represented by the fact that the depletion of Th17 cells was observed in the leukapheresis but not the sigmoid biopsy cohort. This discrepancy can be explained by differences in the technical procedure, age, and duration of ART, as well as the health status and composition of the CCR6+ T-cell pool (Th17, Tregs, etc.) in blood versus colon. Indeed, in the sigmoid biopsy cohort HIV− controls were older compared with the HIV+ individuals (median: 60 versus 55-year old, Table 1) and were included based on their need for cancer screening, with one being positive for hepatitis A and B viruses. Nevertheless, studies by other groups demonstrated the normalization of CD4+ T cell and Th17/Th22 frequencies in the sigmoid biopsies of HIV+ individuals upon long-term ART compared with HIV− individuals [54,99,100]. Consistently, HIV+ individuals included in the sigmoid biopsy cohort received ART for a longer period of time compared with those in the leukapheresis cohorts (median: 72 versus 50 months), thus, suggesting a potential restoration of CCR6+ T cells in the colon and blood upon long-term ART. Finally, whether the contribution of CCR6+ versus CCR6− T cells to HIV reservoirs differ at distinct mucosal sites such as ileum versus colon requires further investigations.
In conclusion, our findings identified CCR6 as a marker for CD4+ T cells enriched in HIV reservoirs in both colon and blood and emphasize the need for novel targeted cell-specific viral eradication strategies. To date, the only demonstrably curative intervention for HIV is bone marrow transplant using a donor homozygous for the CCR5/Δ32 deletion [101]. However, such strategies cannot be applied at large scale. Our current findings, together with those published by other groups [64,65], point to the potential beneficial effect of a temporary depletion of CD4+CCR6+ T cells, depletion that may result in a significant reduction of HIV reservoirs in ART-treated individuals. Although the antibody-mediated depletion of B cells is beneficial for autoimmunity [102], the success of CCR6+CD4+ T-cell depletion in HIV-infected individuals will require careful investigations in animal models and clinical trials. Alternatively, the recent identification of positive/negative regulators of HIV replication preferentially expressed in Th17 cells [45,47] open the path for developing new Th17-specific anti-HIV therapies.
Acknowledgements
The authors acknowledge the contribution of Dr Dominique Gauchat (Flow Cytometry Core Facility, CHUM-Research Centre, Montreal, Québec, Canada) for expert technical support with flow cytometry analysis and sorting, Mr Mario Legault for help with ethical approvals and informed consents, and Josée Girouard and Angie Massicotte for their critical contribution to sigmoid biopsies, blood, and leukapheresis collection from HIV-infected and uninfected donors. Finally, the authors acknowledge human donors for their gift of biological samples essential for this study.
A.G. and T.R.W.S. performed the majority of the experiments, analyzed the results, prepared the figures, and contributed to manuscript writing. D.P., V.S.W., and Y.Z. contributed to leukapheresis preparation, cell sorting by magnetic-activated cell sorting and FACS, and HIV DNA quantification. R.F. and N.C. provided protocols for HIV DNA quantification. V.M. and M.P.G. contributed to sigmoid biopsy and blood collection. E.A.C. and B.S. provided experimental protocols and contributed to manuscript writing. J-P.R. was involved in study volunteer recruitment, access to clinical information, study design, and manuscript writing. P.A. designed the study, analyzed results, designed the figures, and wrote the manuscript. All authors read and approved the manuscript.
The study was supported by grants from the Canadian Institutes of Health Research (CIHR) (#MOP-82849; #MOP-114957) to P.A., grants from the CIHR Canadian HIV Trials Network (CTN #247), the Fonds de Recherche du Québec-Santé (FRQ-S)/AIDS and Infectious Diseases Network, Québec, Canada to J-P.R., and by The Canadian HIV Cure Enterprise Team Grant HIG-133050 (to P.A., E.A.C., and J-P.R.) from the CIHR in partnership with CANFAR and IAS. B.S. received support from NIH-NIAID R01 AI057020. J-P.R. holds a Louis Lowenstein Chair in Hematology and Oncology, McGill University. The funding institutions played no role in the design, collection, analysis, and interpretation of data.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Annie Gosselin and Tomas Raul Wiche Salinas contributed equally to the generation of the results.
Jean-Pierre Routy and Petronela Ancuta contributed equally to the design of the study.
References
- 1.Barre-Sinoussi F, Ross AL, Delfraissy JF. Past, present and future: 30 years of HIV research. Nat Rev Microbiol 2013; 11:877–883. [DOI] [PubMed] [Google Scholar]
- 2.Katlama C, Deeks SG, Autran B, Martinez-Picado J, van Lunzen J, Rouzioux C, et al. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet 2013; 381:2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin AR, Siliciano RF. Progress toward HIV eradication: case reports, current efforts, and the challenges associated with cure. Annu Rev Med 2016; 67:215–228. [DOI] [PubMed] [Google Scholar]
- 4.Schnittman SM, Psallidopoulos MC, Lane HC, Thompson L, Baseler M, Massari F, et al. The reservoir for HIV-1 in human peripheral blood is a T cell that maintains expression of CD4. Science 1989; 245:305–308. [DOI] [PubMed] [Google Scholar]
- 5.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1995; 1:1284–1290. [DOI] [PubMed] [Google Scholar]
- 6.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997; 94:13193–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387:183–188. [DOI] [PubMed] [Google Scholar]
- 8.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999; 5:512–517. [DOI] [PubMed] [Google Scholar]
- 9.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278:1291–1295. [DOI] [PubMed] [Google Scholar]
- 10.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–1300. [DOI] [PubMed] [Google Scholar]
- 11.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9:727–728. [DOI] [PubMed] [Google Scholar]
- 12.Davey RT, Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A 1999; 96:15109–15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun TW, Davey RT, Jr, Engel D, Lane HC, Fauci AS. Re-emergence of HIV after stopping therapy. Nature 1999; 401:874–875. [DOI] [PubMed] [Google Scholar]
- 14.Bruner KM, Hosmane NN, Siliciano RF. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol 2015; 23:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 2012; 37:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun TW, Moir S, Fauci AS. HIV reservoirs as obstacles and opportunities for an HIV cure. Nat Immunol 2015; 16:584–589. [DOI] [PubMed] [Google Scholar]
- 17.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: what, where and how to target them. Nat Rev Microbiol 2016; 14:55–60. [DOI] [PubMed] [Google Scholar]
- 18.Hong FF, Mellors JW. Changes in HIV reservoirs during long-term antiretroviral therapy. Curr Opin HIV AIDS 2015; 10:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 2014; 111:2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016; 530:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed K, Parissenti AM. The effect of ABCB1 genetic variants on chemotherapy response in HIV and cancer treatment. Pharmacogenomics 2011; 12:1465–1483. [DOI] [PubMed] [Google Scholar]
- 22.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 2012; 12:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M, Hosmane NN, Bullen CK, Capoferri A, Yang HC, Siliciano JD, et al. A primary CD4(+) T cell model of HIV-1 latency established after activation through the T cell receptor and subsequent return to quiescence. Nat Protoc 2014; 9:2755–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 2014; 20:425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen TA, Lewin SR. Shocking HIV out of hiding: where are we with clinical trials of latency reversing agents?. Curr Opin HIV AIDS 2016; 11:394–401. [DOI] [PubMed] [Google Scholar]
- 26.Chomont N, Dafonseca S, Vandergeeten C, Ancuta P, Sekaly RP. Maintenance of CD4+ T-cell memory and HIV persistence: keeping memory, keeping HIV. Curr Opin HIV AIDS 2011; 6:30–36. [DOI] [PubMed] [Google Scholar]
- 27.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Masson A, Kirilovsky A, Zoorob R, Avettand-Fenoel V, Morin V, Oudin A, et al. Blimp-1 overexpression is associated with low HIV-1 reservoir and transcription levels in central memory CD4+ T cells from elite controllers. AIDS 2014; 28:1567–1577. [DOI] [PubMed] [Google Scholar]
- 29.Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, et al. Low levels of SIV infection in sooty mangabey central memory CD(4)(+) T cells are associated with limited CCR5 expression. Nat Med 2011; 17:830–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 2014; 20:139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, et al. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol 2014; 88:14070–14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 2013; 210:143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, et al. PD-1+ and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 2016; 22:754–761. [DOI] [PubMed] [Google Scholar]
- 34.Jaafoura S, de Goer de Herve MG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4(+) memory T ells. Nat Commun 2014; 5:5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeshita M, Suzuki K, Kassai Y, Takiguchi M, Nakayama Y, Otomo Y, et al. Polarization diversity of human CD4(+) stem cell memory T cells. Clin Immunol 2015; 159:107–117. [DOI] [PubMed] [Google Scholar]
- 36.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011; 34:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sallusto F. Heterogeneity of human CD4(+) T cells against microbes. Annu Rev Immunol 2016; 34:317–334. [DOI] [PubMed] [Google Scholar]
- 38.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol 2008; 8:337–348. [DOI] [PubMed] [Google Scholar]
- 39.Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol 2013; 8:477–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenchley JM, Paiardini M. Immunodeficiency lentiviral infections in natural and nonnatural hosts. Blood 2011; 118:847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, et al. Peripheral blood CCR4+ CCR6+ and CXCR3+ CCR6+ CD4+ T cells are highly permissive to HIV-1 infection. J Immunol 2010; 184:1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El Hed A, Khaitan A, Kozhaya L, Manel N, Daskalakis D, Borkowsky W, et al. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J Infect Dis 2010; 201:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, Kared H, et al. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin β7. J Immunol 2011; 186:4618–4630. [DOI] [PubMed] [Google Scholar]
- 44.Alvarez Y, Tuen M, Shen G, Nawaz F, Arthos J, Wolff MJ, et al. Preferential HIV infection of CCR6+ Th17 cells is associated with higher levels of virus receptor expression and lack of CCR5 ligands. J Virol 2013; 87:10843–10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernier A, Cleret-Buhot A, Zhang Y, Goulet JP, Monteiro P, Gosselin A, et al. Transcriptional profiling reveals molecular signatures associated with HIV permissiveness in Th1Th17 cells and identifies peroxisome proliferator-activated receptor gamma as an intrinsic negative regulator of viral replication. Retrovirology 2013; 10:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touzot M, Grandclaudon M, Cappuccio A, Satoh T, Martinez-Cingolani C, Servant N, et al. Combinatorial flexibility of cytokine function during human T helper cell differentiation. Nat Commun 2014; 5:3987. [DOI] [PubMed] [Google Scholar]
- 47.Cleret-Buhot A, Zhang Y, Planas D, Goulet JP, Monteiro P, Gosselin A, et al. Identification of novel HIV-1 dependency factors in primary CCR4CCR6Th17 cells via a genome-wide transcriptional approach. Retrovirology 2015; 12:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christensen-Quick A, Lafferty M, Sun L, Marchionni L, DeVico A, Garzino-Demo A. Human Th17 cells lack HIV-inhibitory RNases and are highly permissive to productive HIV infection. J Virol 2016; 90:7833–7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez-Garcia M, Barr FD, Crist SG, Fahey JV, Wira CR. Phenotype and susceptibility to HIV infection of CD4(+) Th17 cells in the human female reproductive tract. Mucosal Immunol 2014; 7:1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKinnon LR, Nyanga B, Kim CJ, Izulla P, Kwatampora J, Kimani M, et al. Early HIV-1 infection is associated with reduced frequencies of cervical Th17 cells. J Acquir Immune Defic Syndr 2015; 68:6–12. [DOI] [PubMed] [Google Scholar]
- 51.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med 2008; 14:421–428.18376406 [Google Scholar]
- 52.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol 2012; 30:149–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol 2012; 10:655–666. [DOI] [PubMed] [Google Scholar]
- 54.Kim CJ, McKinnon LR, Kovacs C, Kandel G, Huibner S, Chege D, et al. Mucosal Th17 cell function is altered during HIV infection and is an independent predictor of systemic immune activation. J Immunol 2013; 191:2164–2173. [DOI] [PubMed] [Google Scholar]
- 55.Chege D, Sheth PM, Kain T, Kim CJ, Kovacs C, Loutfy M, et al. Sigmoid Th17 populations, the HIV latent reservoir, and microbial translocation in men on long-term antiretroviral therapy. AIDS 2011; 25:741–749. [DOI] [PubMed] [Google Scholar]
- 56.Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol 2013; 21:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ponte R, Mehraj V, Ghali P, Couedel-Courteille A, Cheynier R, Routy JP. Reversing gut damage in HIV infection: using non-human primate models to instruct clinical research. EBioMedicine 2016; 4:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stieh DJ, Matias E, Xu H, Fought AJ, Blanchard JL, Marx PA, et al. Th17 cells are preferentially infected very early after vaginal transmission of SIV in macaques. Cell Host Microbe 2016; 19:529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basu R, Hatton RD, Weaver CT. The Th17 family: flexibility follows function. Immunol Rev 2013; 252:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gagliani N, Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 2015; 523:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, et al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity 2011; 35:972–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muranski P, Restifo NP. Essentials of Th17 cell commitment and plasticity. Blood 2013; 121:2402–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wacleche VS, Goulet JP, Gosselin A, Monteiro P, Soudeyns H, Fromentin R, et al. New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy. Retrovirology 2016; 13:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun H, Kim D, Li X, Kiselinova M, Ouyang Z, Vandekerckhove L, et al. Th1/17 polarization of CD4 T cells supports HIV-1 DNA persistence during antiretroviral therapy. J Virol 2015; 89:11284–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khoury G, Anderson JL, Fromentin R, Hartogensis W, Smith MZ, Bacchetti P, et al. Persistence of integrated HIV DNA in CXCR3 + CCR6 + memory CD4+ T-cells in HIV-infected individuals on antiretroviral therapy. AIDS 2016; 30:1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shacklett BL, Yang O, Hausner MA, Elliott J, Hultin L, Price C, et al. Optimization of methods to assess human mucosal T-cell responses to HIV infection. J Immunol Methods 2003; 279:17–31. [DOI] [PubMed] [Google Scholar]
- 67.Hayes TL, Asmuth DM, Critchfield JW, Knight TH, McLaughlin BE, Yotter T, et al. Impact of highly active antiretroviral therapy initiation on CD4(+) T-cell repopulation in duodenal and rectal mucosa. AIDS 2013; 27:867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boulassel MR, Spurll G, Rouleau D, Tremblay C, Edwardes M, Sekaly RP, et al. Changes in immunological and virological parameters in HIV-1 infected subjects following leukapheresis. J Clin Apher 2003; 18:55–60. [DOI] [PubMed] [Google Scholar]
- 69.Roederer M. Compensation in flow cytometry. Curr Protoc Cytom 2002; Chapter 1:Unit 1.14. [DOI] [PubMed] [Google Scholar]
- 70.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol 2007; 8:639–646. [DOI] [PubMed] [Google Scholar]
- 71.Steinfelder S, Floess S, Engelbert D, Haeringer B, Baron U, Rivino L, et al. Epigenetic modification of the human CCR6 gene is associated with stable CCR6 expression in T cells. Blood 2011; 117:2839–2846. [DOI] [PubMed] [Google Scholar]
- 72.Vandergeeten C, Fromentin R, DaFonseca S, Lawani MB, Sereti I, Lederman MM, et al. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 2013; 121:4321–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hughes SH, Coffin JM. What integration sites tell us about HIV persistence. Cell Host Microbe 2016; 19:588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang C, Kang SG, Lee J, Sun Z, Kim CH. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol 2009; 2:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loiseau C, Requena M, Mavigner M, Cazabat M, Carrere N, Suc B, et al. CCR6 regulatory T cells blunt the restoration of gut Th17 cells along the CCR6-CCL20 axis in treated HIV-1-infected individuals. Mucosal Immunol 2016; 9:1137–1150. [DOI] [PubMed] [Google Scholar]
- 76.Wong MT, Ong DE, Lim FS, Teng KW, McGovern N, Narayanan S, et al. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 2016; 45:442–456. [DOI] [PubMed] [Google Scholar]
- 77.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, et al. Phenotypic and functional features of human Th17 cells. J Exp Med 2007; 204:1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wan Q, Kozhaya L, ElHed A, Ramesh R, Carlson TJ, Djuretic IM, et al. Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6+ human memory T cells. J Exp Med 2011; 208:1875–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 2015; 163:381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mavigner M, Cazabat M, Dubois M, L’Faqihi FE, Requena M, Pasquier C, et al. Altered CD4+ T cell homing to the gut impairs mucosal immune reconstitution in treated HIV-infected individuals. J Clin Invest 2012; 122:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DaFonseca S, Niessl J, Pouvreau S, Wacleche VS, Gosselin A, Cleret-Buhot A, et al. Impaired Th17 polarization of phenotypically naive CD4(+) T-cells during chronic HIV-1 infection and potential restoration with early ART. Retrovirology 2015; 12:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mercer F, Khaitan A, Kozhaya L, Aberg JA, Unutmaz D. Differentiation of IL-17-producing effector and regulatory human T cells from lineage-committed naive precursors. J Immunol 2014; 193:1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, et al. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol 2012; 5:646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Micci L, Cervasi B, Ende ZS, Iriele RI, Reyes-Aviles E, Vinton C, et al. Paucity of IL-21-producing CD4(+) T cells is associated with Th17 cell depletion in SIV infection of rhesus macaques. Blood 2012; 120:3925–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schuetz A, Deleage C, Sereti I, Rerknimitr R, Phanuphak N, Phuang-Ngern Y, et al. Initiation of ART during early acute hiv infection preserves mucosal Th17 function and reverses HIV-related immune activation. PLoS Pathog 2014; 10:e1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.d’Ettorre G, Baroncelli S, Micci L, Ceccarelli G, Andreotti M, Sharma P, et al. Reconstitution of intestinal CD4 and Th17 T cells in antiretroviral therapy suppressed HIV-infected subjects: implication for residual immune activation from the results of a clinical trial. PLoS One 2014; 9:e109791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kok A, Hocqueloux L, Hocini H, Carriere M, Lefrou L, Guguin A, et al. Early initiation of combined antiretroviral therapy preserves immune function in the gut of HIV-infected patients. Mucosal Immunol 2015; 8:127–140. [DOI] [PubMed] [Google Scholar]
- 88.Dillon SM, Lee EJ, Donovan AM, Guo K, Harper MS, Frank DN, et al. Enhancement of HIV-1 infection and intestinal CD4+ T cell depletion ex vivo by gut microbes altered during chronic HIV-1 infection. Retrovirology 2016; 13:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cameron PU, Saleh S, Sallmann G, Solomon A, Wightman F, Evans VA, et al. Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc Natl Acad Sci U S A 2010; 107:16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature 2009; 458:1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu H, Nau M, Ehrenberg P, Chenine AL, Macedo C, Zhou Y, et al. Distinct gene-expression profiles associated with the susceptibility of pathogen-specific CD4 T cells to HIV-1 infection. Blood 2013; 121:1136–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee MO, Hobbs PD, Zhang XK, Dawson MI, Pfahl M. A synthetic retinoid antagonist inhibits the human immunodeficiency virus type 1 promoter. Proc Natl Acad Sci U S A 1994; 91:5632–5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li P, Kaiser P, Lampiris HW, Kim P, Yukl SA, Havlir DV, et al. Stimulating the RIG-I pathway to kill cells in the latent HIV reservoir following viral reactivation. Nat Med 2016; 22:807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Duhen T, Duhen R, Lanzavecchia A, Sallusto F, Campbell DJ. Functionally distinct subsets of human FOXP3+ Treg cells that phenotypically mirror effector Th cells. Blood 2012; 119:4430–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seddiki N, Santner-Nanan B, Martinson J, Zaunders J, Sasson S, Landay A, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med 2006; 203:1693–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Favre D, Lederer S, Kanwar B, Ma ZM, Proll S, Kasakow Z, et al. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog 2009; 5:e1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moreno-Fernandez ME, Zapata W, Blackard JT, Franchini G, Chougnet CA. Human regulatory T cells are targets for human immunodeficiency Virus (HIV) infection, and their susceptibility differs depending on the HIV type 1 strain. J Virol 2009; 83:12925–12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bin Dhuban K, d’Hennezel E, Nashi E, Bar-Or A, Rieder S, Shevach EM, et al. Coexpression of TIGIT and FCRL3 identifies Helios+ human memory regulatory T cells. J Immunol 2015; 194:3687–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Macal M, Sankaran S, Chun TW, Reay E, Flamm J, Prindiville TJ, et al. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol 2008; 1:475–488. [DOI] [PubMed] [Google Scholar]
- 100.Sheth PM, Chege D, Shin LY, Huibner S, Yue FY, Loutfy M, et al. Immune reconstitution in the sigmoid colon after long-term HIV therapy. Mucosal Immunol 2008; 1:382–388. [DOI] [PubMed] [Google Scholar]
- 101.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 2009; 360:692–698. [DOI] [PubMed] [Google Scholar]
- 102.Marquez AC, Horwitz MS. The role of latently infected B cells in CNS autoimmunity. Front Immunol 2015; 6:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


