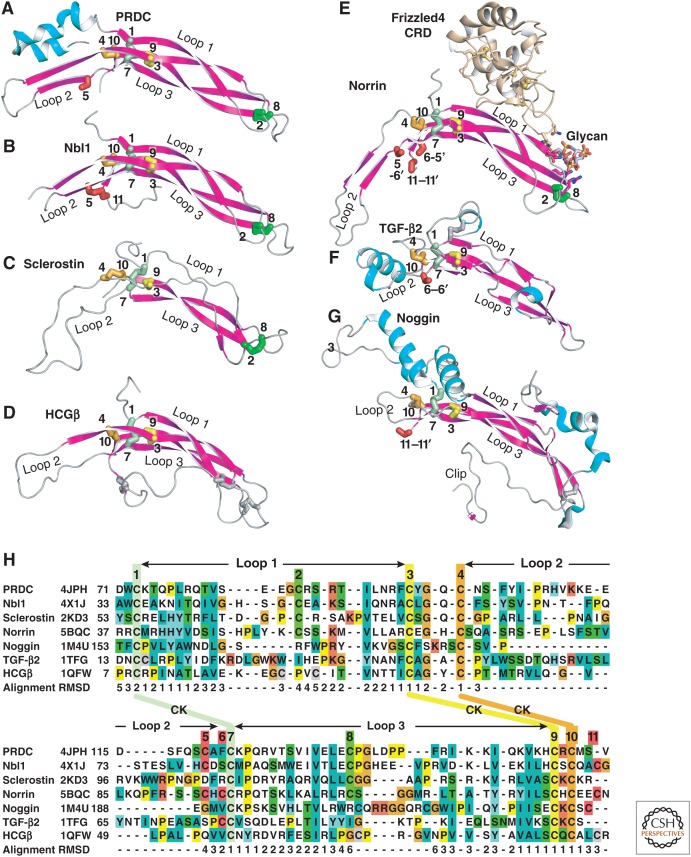
Figure 11.
Structural alignment of Dan family monomers to cystine knot (CK) growth factor (CKGF) superfamily representatives with similar monomer–monomer orientations. (A–G) CKGF monomers in cartoon with α-helices in cyan and β-strands in magenta. Disulfides are shown in stick and are color-coded with their cysteines numbered as in panel H. Cysteines in intermolecular disulfides are indicated by showing the partner Cys in the other monomer with its number preceded by a dash and followed by a hyphen. All monomers but sclerostin are from dimeric structures. A frizzled domain and glycan cocrystallized with norrin are shown in cartoon and stick, respectively. Monomers are in identical orientations, and aligned in two columns. (H) Structure-based sequence alignment. Cysteines disulfide-bonded to one another are in identical colors, except those that participate in interchain disulfides in some families and intrachain disulfides in others (positions 5, 6, and 11) are shown in red. Disulfides unique to particular families are shown in thinner silver stick and colored silver in panel H. Other residues are colored according to ClustalX. CK disulfides are labeled between the two rows of the alignment and link Cys residues distant in sequence; the three long loops between CK disulfides are also marked. For alignments to CKGF families with monomers dimerized in different orientations (neurotrophin in the nerve growth factor [NGF] family, and platelet-derived growth factor [PDGF] in the PDGF family) (see Zhou and Springer 2014). CKGF and CTCK monomers (PDB codes are shown in panel H) were aligned to one another structurally using DeepAlign (Wang et al. 2013). Structurally equivalent residues are shown aligned by sequence as described in Figure 5 legend. Cα atom root mean square deviation (RMSD, Å) of monomers in each position relative to the average at each position are shown below the alignment. Gaps were closed to minimize the length of the alignment; corresponding positions lack an RMSD value.