Figure 2.
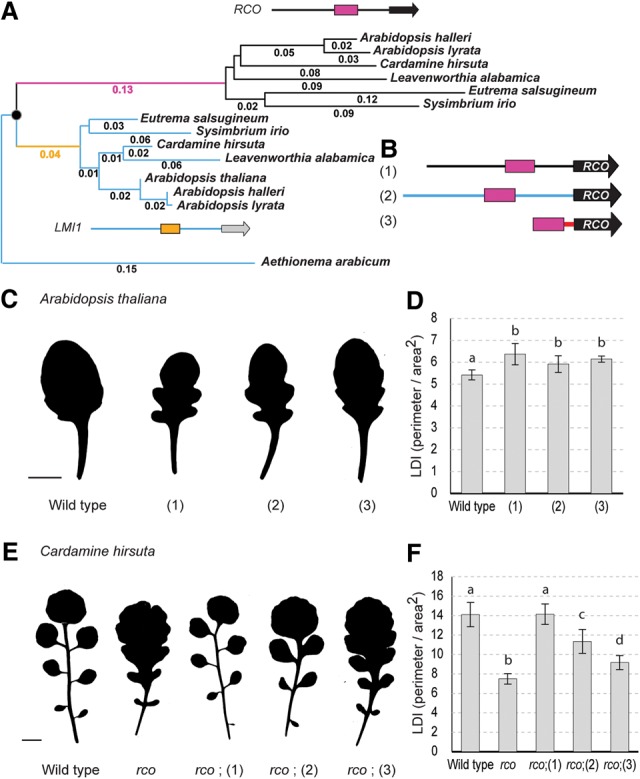
ChRCOenh500 evolved nonneutrally and is sufficient to increase leaf complexity when driving RCO in A. thaliana and C. hirsuta. (A) The RCO enhancer (magenta; depicted on a black gene model) experienced an increased base substitution rate (likelihood ratio test: ΔlnL = 9.64; P < 0.005) compared with the LMI1 enhancer (yellow; depicted on a blue gene model), consistent with positive selection (see the Supplemental Material for detailed analysis). Base substitution rates are indicated next to individual branches. (B) Constructs (1–3) used to express RCO in the RCO domain contain the upstream sequence of ChLMI1 (blue line), ChRCO (black line), or the CaMV 35S minimal promoter (red line) with enhancer BRCO (magenta). (C,E) Representative leaf 8 silhouettes from A. thaliana (C) and C. hirsuta (E) wild-type and transgenic plants. Bar, 1 cm. (D,F) Leaf dissection index ([LDI] perimeter/area2) calculated from A. thaliana (D) and C. hirsuta (F) leaf 8 silhouettes. Graphs indicate average LDI and standard deviation. Letters indicate significant differences between groups as indicated by ANOVA and post-hoc Tukey's test. P < 0.01. For constructs 2 and 3, at least three independent T2 lines were analyzed with n > 12.
