Figure 6.
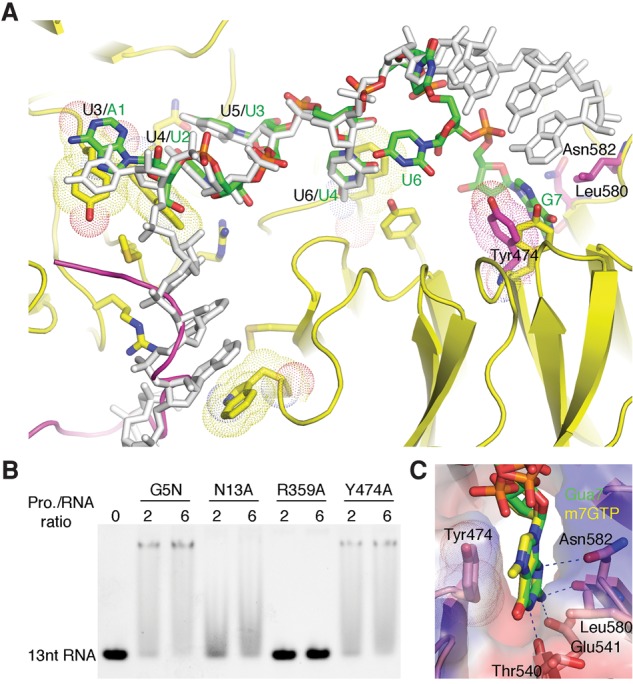
Alternative RNA-binding mode of G5N. (A) The soaked-in 7-mer Sm RNA is superimposed with the cocrystal structure with 13-mer RNA. The 7-mer RNA is shown in a stick model colored according to the scheme in Figure 3, while the cocrystallized 13-mer RNA is colored gray. The green labels indicate the identity of the 7-mer RNA nucleotides, while the preceding black labels separated by a slash indicate nucleotide positions in the cocrystallized 13-mer RNA. Tyr474, Asn582, and Leu580 from the soaked-in G5N structure are superimposed as a stick model, with carbon atoms colored magenta. Please note that the conformation of Tyr474 is changed from that in the cocrystallized structure (yellow) to accommodate the binding of Gua7. A loop connecting blades 5 and 6 of the first WD40 domain, which is ordered in native and soaked-in G5N structures (magenta) but disordered in the cocrystal structure, is superimposed. (B) EMSA analysis of the binding of the Y474A mutant to the 13-mer RNA. Wild-type G5N and N13A and R359A mutants were used as controls. (C) Binding of the m7GTP cap analog to G5N at the site that bound Gua7 in the soaked-in 7-mer RNA complex.
