Figure 7.
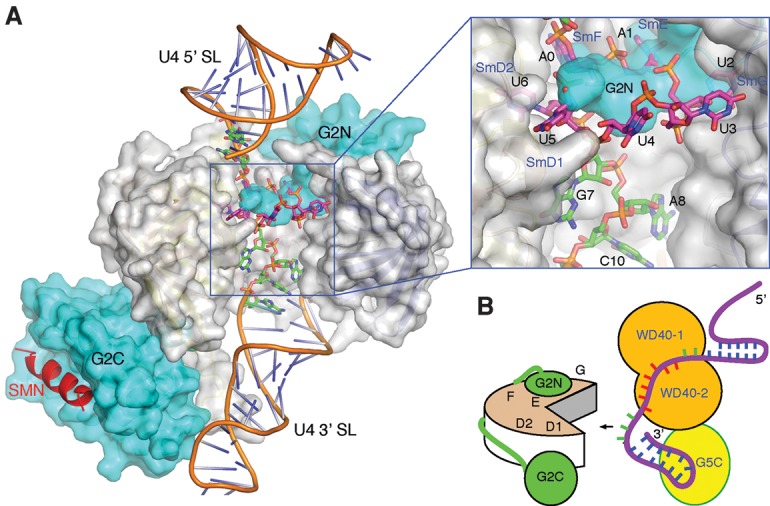
A stretch of nucleotides 3′ to the Sm site is important for snRNP assembly. (A) A model with Gemin2 and a short region of SMN (Protein Data Bank [PDB] ID 3s6n) bound to the U4 snRNA (PDB ID 4wzj). The SmB/D3 heterodimer was removed for viewing clarity and to mimick the pentameric Sm assembly intermediate. Gemin2 and SmD1–D2–F–E–G are shown in a surface representation, with Gemin2 colored in cyan, and the Sm proteins colored in gray. The SMN fragment is shown in a cartoon representation colored in red. The 13 nt used for cocrystallization is shown in a stick representation, with the carbon bonds colored magenta for nucleotides belonging to the heptameric A0A1U2U3U4U5U6 sequence involved in G5N binding and green for the remaining nucleotides. U4 snRNA outside of the 13-mer region is shown in a cartoon representation with the backbone colored brown. An inset at the top right displays an enlarged view of the boxed area of the main figure. (B) An illustration of how Gemin5 may delimit the free single-stranded region of U4 snRNA 3′ to the AAUUUUU G5N recognition sequence (colored red) for presentation to Gemin2 (labeled G2N and G2C) bound the Sm pentamer. A hypothetical C-terminal region of Gemin5 bound to the 3′ stem–loop is shown.
