Figure 3.
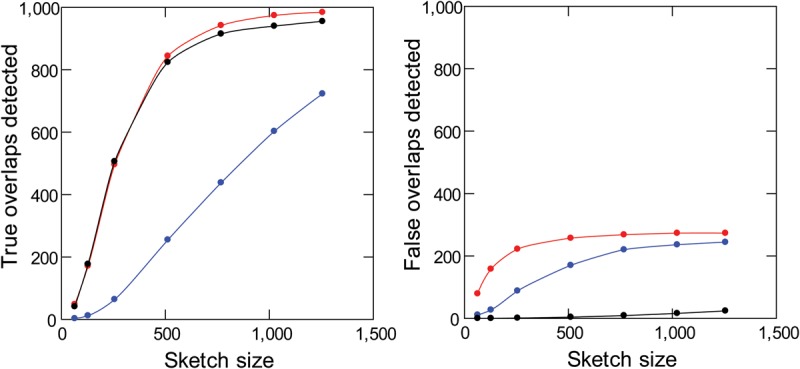
Sensitivity and specificity of read overlap detection with masking of repetitive k-mers. Simulated PacBio reads from D. melanogaster (1000 pairs of 10-kb sequences with 2-kb overlaps) were subjected to standard MHAP (blue), MHAP with masking of low-frequency k-mers (red), or MHAP with masking of low-frequency and high-frequency k-mers (black). Note that masking of low- and high-frequency k-mers cause a huge improvement in specificity (right) with minimal losses in sensitivity (left). The reference list of valid k-mers came from Illumina reads.
