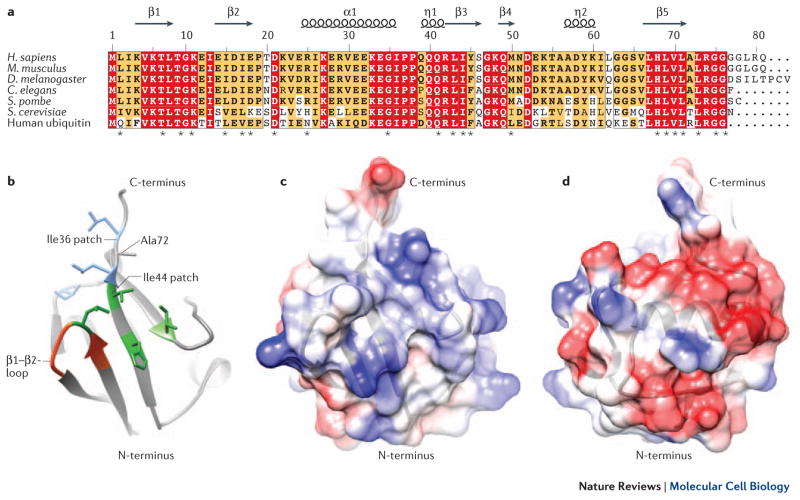
Figure 1. NEDD8 Structure.
A Structure-based multiple sequence alignment of NEDD8 from indicated species and human ubiquitin, highlighting identical residues (red) and similar residues (yellow). Asterisks signify residues found to be essential for NEDD8 function in S. pombe63. B Structural representation of the NEDD8 hydrophobic patches, contributing most of the known interfaces for interaction with binding partners (Table 1). The side chains contributing to the Ile36 patch are shown in blue, the residues of the Ile44 patch in green and the β1/β2-loop in red. Those interfaces are conserved in ubiquitin. Ala72, which is responsible for discrimination between NEDD8 and ubiquitin by the respective E1 enzymes, is indicated. C, D Structural representations of the NEDD8-specific charged surface patches. Acidic patches are depicted in red and basic surfaces in blue. These surfaces might be responsible for interactions that discriminate between NEDD8 and ubiquitin.