Figure 2. PpSMF1 and PpSCRM1 are required for stomatal development in the moss Physcomitrella patens.
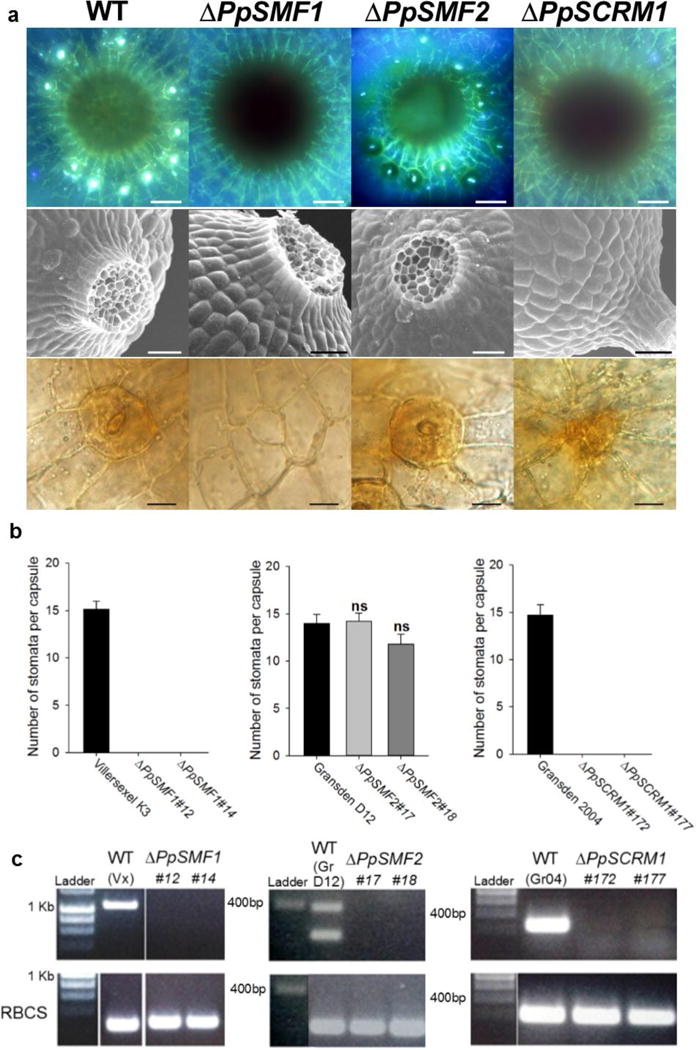
(a) Stacked UV fluorescence images (upper panel), scanning electron microscope images (middle panel) and bright field images (bottom panel) showing the spore capsule base and epidermal close-ups from P. patens wild-type, ΔPpSMF1, ΔPpSMF2 and ΔPpSCRM1 knock-out mutants, respectively. The top panel wild-type representative is from Villersexel K3 ecotype of P. patens, the middle panel wild-type representative is from the Gransden D12 ecotype and the bottom panel wild-type relates to the Gransden 2004 ecotype. There were no discernible differences between the sporophytes of the different background lines. For both of the ΔPpSCRM1 lines generated we observed one such instance of aborted stomata (see bottom right panel) in the 7 capsules of each line surveyed. (b) Number of stomata formed per sporophyte in two independent lines of each genotype versus wild-type controls. Error bars indicate one standard error of the mean. For ΔPpSMF1 and ΔPpSCRM1 and the corresponding wild-types, n = 7 capsules of each line were analysed. For ΔPpSMF2 and wild-type background, 5 capsules were surveyed. A One-way ANOVA was performed to test for differences between the wild-type and ΔPpSMF2 lines and no significant differences (denoted ns) were found. (c) RT-PCR to confirm loss of the respective transcript in each of the P. patens knock-out lines (top panel). A Rubisco (RBCS) control was run to verify the integrity of the produced cDNA (Bottom panel). For labelling purposes the wild-types Villersexel K3, Gransden D12 and Gransden 2004 are denoted Vx, GrD12 and Gr04. For PpSMF2 two bands were amplified in the control for which the smaller 239bp product represents the size expected for PpSMF2. Scale bars in a = 50 μm in the top and middle panels, in the bottom panel = 15 μm.
