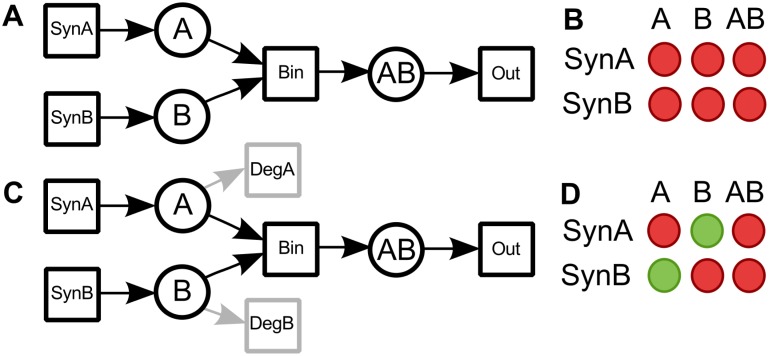
Fig 1. In silico knockout matrix and the corresponding PN.
(A) A small PN, consisting of three places, protein A, protein B, and a protein complex AB, and four transitions, SynA, SynB, Bin, and Out, which describe the synthesis of A, the synthesis of B, the binding of both proteins, and the outflow of the complex to the environment, respectively. (B) The in silico knockout matrix of the PN shown in part A. The matrix has a row for each input transition, SynA and SynB, and columns for each substance, A, B, and AB. The binary values of an entry are color-coded by either a red or a green circle. An entry becomes red, if the corresponding place is not the outgoing place of a transition that is part of a still functional T-invariant. The entry is green, if the corresponding place is still the output place of a transition that is part of a T-invariant. For the PN in part A, the knockout of either SynA or SynB is sufficient to have a negative effect on all substances, A, B, and AB. Consequently, all entries in the matrix are red. (C) The PN is modified by adding the output transitions, DegA and DegB. Now, the knockout analysis becomes more specific. The knockout of SynA blocks the production of A and complex AB, but B is not affected. Vice versa, the knockout of SynB leaves A unaffected. (D) The corresponding in silico knockout matrix of the modified PN shown in part C.