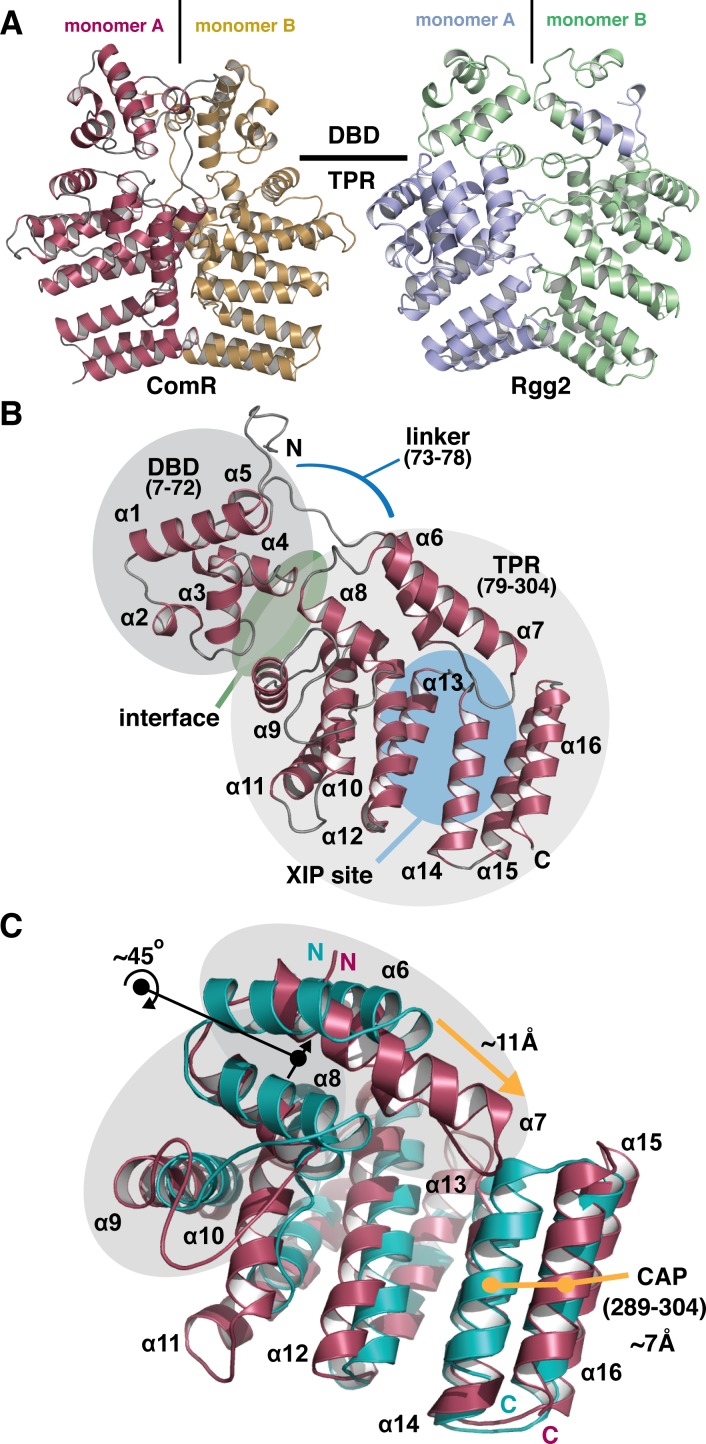
Fig 4. Overall structure of apo-ComR from Streptococcus suis.
(A) Cartoon representation of the asymmetric unit of the native crystal form compared to the biological unit of Rgg2 (PDB code 4YV6). Each monomer is colored and the approximate boundary between the DNA binding domain (DBD) and the tetratricopeptide repeat domain (TPR) is indicated. (B) The biological unit of the apo-form of ComR. The DBD domain and TPR domains are shaded in grey with primary sequence boundaries listed in parenthesis with the extended loop that links the domains indicated by a blue line. The XIP binding region in the TPR is highlighted with a blue oval, in addition to the interface region between the DBD and TPR with a green oval. Each secondary structure element is labeled in by number order from N to C terminus (α = α-helix) (C) Alignment of the S. suis TRP (red) and Rgg2 TPR (teal). Black arrows and dots indicate a rotation of α-helices and orange arrows and dots show a translation of α-helices. The regions shaded in grey and the C-terminal CAP helix show significant differences.