Abstract
Background
Left atrial enlargement in mitral regurgitation (MR) predicts a poor prognosis. The regulatory mechanisms of atrial myocyte hypertrophy of MR patients remain unknown.
Methods and Results
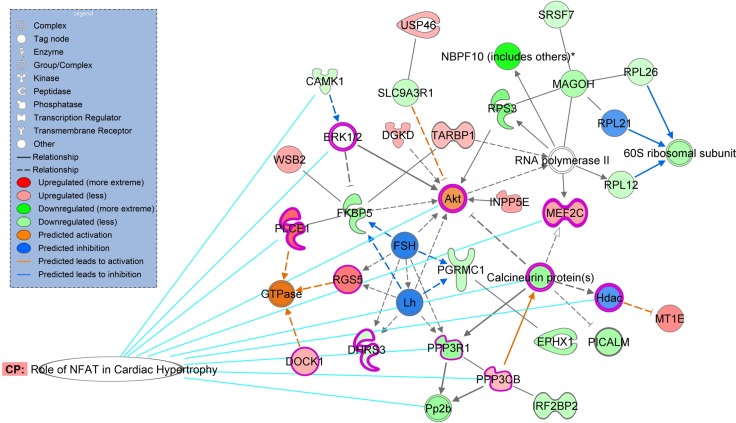
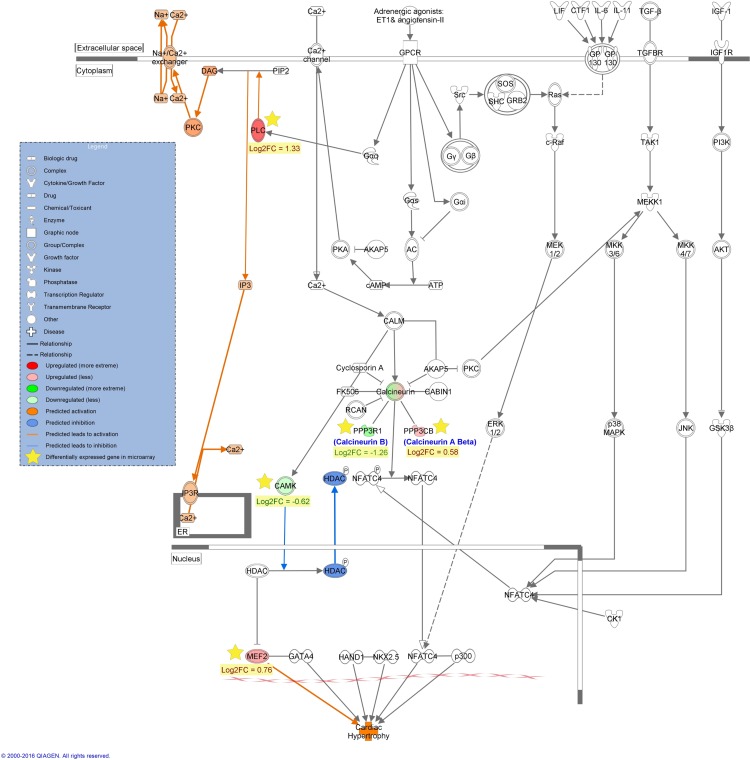
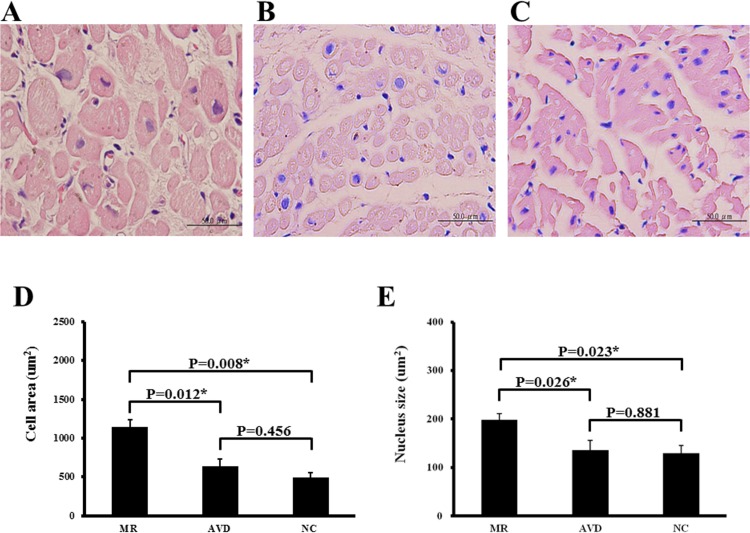
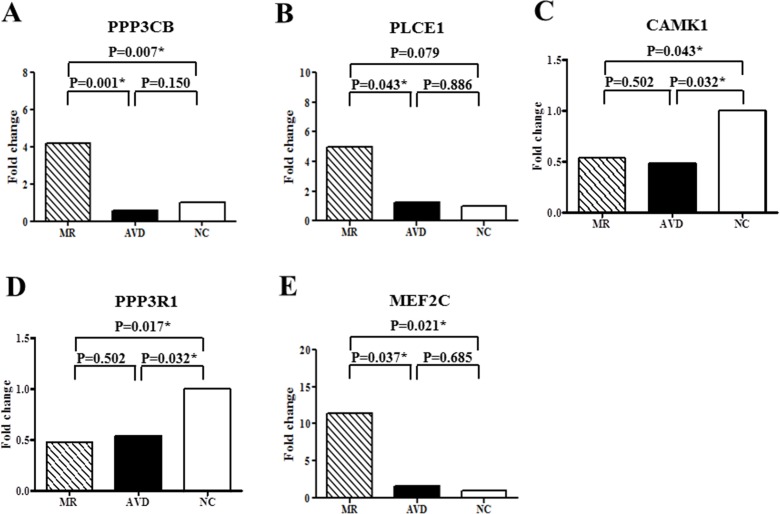
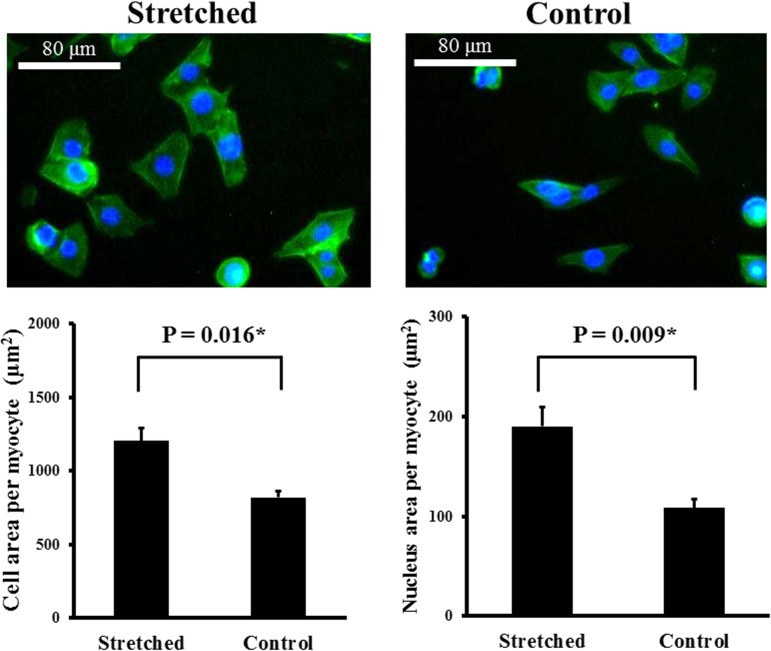
This study comprised 14 patients with MR, 7 patients with aortic valve disease (AVD), and 6 purchased samples from normal subjects (NC). We used microarrays, enrichment analysis and quantitative RT-PCR to study the gene expression profiles in the left atria. Microarray results showed that 112 genes were differentially up-regulated and 132 genes were differentially down-regulated in the left atria between MR patients and NC. Enrichment analysis of differentially expressed genes demonstrated that “NFAT in cardiac hypertrophy” pathway was not only one of the significant associated canonical pathways, but also the only one predicted with a non-zero score of 1.34 (i.e. activated) through Ingenuity Pathway Analysis molecule activity predictor. Ingenuity Pathway Analysis Global Molecular Network analysis exhibited that the highest score network also showed high association with cardiac related pathways and functions. Therefore, 5 NFAT associated genes (PPP3R1, PPP3CB, CAMK1, MEF2C, PLCE1) were studies for validation. The mRNA expressions of PPP3CB and MEF2C were significantly up-regulated, and CAMK1 and PPP3R1 were significantly down-regulated in MR patients compared to NC. Moreover, MR patients had significantly increased mRNA levels of PPP3CB, MEF2C and PLCE1 compared to AVD patients. The atrial myocyte size of MR patients significantly exceeded that of the AVD patients and NC.
Conclusions
Differentially expressed genes in the “NFAT in cardiac hypertrophy” pathway may play a critical role in the atrial myocyte hypertrophy of MR patients.
Introduction
Mitral regurgitation (MR) is an important cause of heart failure related to valvular heart disease [1]. Left atrial enlargement has prognostic significance in MR patients undergoing mitral valve surgery [2]. Structural remodeling associated with atrial enlargement, especially pathological hypertrophy of myocytes, developed in the left atrial myocardium of patients with MR [3,4]. However, the molecular regulatory mechanisms and functional biological pathways related to the left atrial myocyte hypertrophy of MR patients remain unclear.
In this study, we aimed to systemically explore the crucial differences in the RNA expression pattern between the left atrial myocardium of MR patients and normal subjects, and the molecular regulatory mechanisms and functional biological pathways related to the atrial myocyte hypertrophy using high-density oligonucleotide microarrays and enrichment analysis. The left atrial myocardium of patients with severe aortic valve disease was also used as a reference for gene analysis of the significant pathways as the left atrial size was smaller in patients with aortic valve disease compared to MR patients. The results of this study may recognize some of the differentially expressed genes and related pathways that contribute to the left atrial myocyte hypertrophy in patients with MR.
Methods
Patient Population
This study enrolled 14 patients with symptomatic severe non-ischemic MR in sinus rhythm (age: 58±9 years), and 7 age-matched patients with symptomatic severe aortic valve disease in sinus rhythm (age: 63±7 years; aortic stenosis in 1, aortic regurgitation in 4, combined aortic stenoregurgitation in 2). Exclusion factors include previous myocardial infarction, febrile disorder, infectious or inflammatory disease, autoimmune disease, malignancy, acute or chronic viral hepatitis or use of immunosuppressive drugs. Written informed consent was obtained from each study patient, and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Institutional Review Board of Chang Gung Memorial Hospital (100-0067C).
Six normal adult left atrial tissue samples (24-year-old Caucasian male, 27-year-old Caucasian male, 30-year-old Asian male, 60-year-old Caucasian female, 76-year-old Caucasian female and 77-year-old Caucasian male) were purchased from BioChain, Newark, CA, USA, and these 6 normal atrial tissues were used as the normal controls for gene analysis.
Specimen Storage
Atrial tissues of non-ischemic MR patients and aortic valve disease patients with heart failure were sampled from the left atrial free wall during surgery. After excision, some atrial tissues were immediately frozen in liquid nitrogen and stored at –80 Celsius, and some were immediately fixed in 3.7% buffered formalin, then embedded in paraffin, and stored until later study for hematoxylin/eosin staining.
Microarray Analysis and Data Processing
RNAs were extracted from the myocardial samples by using a RiboPureTM kit (Ambion, Grand Island, NY, USA) according to the manufacturer's protocol. RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc, Santa Clara, CA, USA). Samples with optical density ratio 260/280 > 1.8 and RNA integrity number > 7.0 were selected and sent for microarray processing. Two hundred fifty ng of total RNA per sample was used for cRNA production by the RiboPureTMRNA extraction kit (Ambion, Grand Island, NY, USA). The quality of cRNA was evaluated using the RNA 6000 pico kit (Agilent Technologies, Santa clara,CA,USA) and the Experion automated electrophoresis station (Bio-Rad Laboratories, Inc., Hercules, CA, USA). A total of 750 ng cRNA was used for hybridization to a human HT12-v4 Illumina Beadchip gene expression array (Illumina, San Diego, CA, USA), including 47231 probes and 28688 annotated genes, according to the manufacturer’s protocol. The arrays were scanned and fluorescence signals obtained using the Illumina Bead Array Reader (Illumina, San Diego, CA, USA). Microarray quality control and normalization was performed using Illumina GenomeStudio data analysis software. The expression level of a gene was represented by the average probe intensity. Functional classes were assigned to all known genes using information from the Gene Ontology database available at the website (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi). Additionally, we applied the activation z-score analysis method [5] to measure activation states (increased or decreased) of the pathways affected by differentially expressed genes. The sign of the calculated z-score will reflect the overall predicted activation state of the biological function (<0: decreased, >0: increased).
Quantitative Determination of RNAs by Real-Time RT-PCR
The RNA samples were quantified using a spectrophotometer. First-strand cDNAs were synthesized with reverse transcriptase and oligo (dT) primers. Real-time quantitative PCR was performed on the ABI Prism 7500 FAST sequence detection system (Applied Biosystems, CA, USA), using SYBR Green PCR Master Mix (Applied Biosystems, CA, USA). The results of RNAs were normalized against 18S gene expression (the endogenous control). The selected genes and primer sequences are presented in Table 1. The microRNAs (miRs) were extracted from the tissues by using a RNA MiniPrep kit (Zymo Research, CA, USA) according to the manufacturer’s protocol. Reverse transcription of miRs was performed using the TaqMan™ microRNA reverse transcription kit (Applied Biosystems, CA, USA) according to manufacturer's recommendations. Briefly, 5 ng of miR was combined with deoxyribonucleoside triphosphates, MultiScribe™ reverse transcriptase, and the primer specific for the target miR (Applied Biosystems, CA, USA). The cDNA was combined with the TaqMan™ assay specific for the target miR. The results of miRs were normalized against U6 snRNA (Applied Biosystems, CA, USA). Quantitative RT-PCR values were presented in △Cq units.
Table 1. Primer Sequences for Real-Time PCR.
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| Human | ||
| PPP3CB | TGGTGGACTTTCACCAGAAAT | GCAGGTGGCTCTTTGAATCT |
| PLCE1 | CTGCGGAAACAGTACGTCAG | CAAAGTTGGGCCTTCATACC |
| CAMK1 | AAGGCAGCATGGAGAATGAG | GCTACAATGTTGGGGTGCTT |
| PPP3R1 | TTATAATCCCAGCCAGTGGTTT | AAAGGCTAGTTCCCCCTTGA |
| MEF2C | AAGGTATCCATGGAACATGAAAG | TGAGTGTGTATATTTTCAGGGATGTT |
| 18S | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
| Mouse | ||
| CAMK1 | AAGCAGGCGGAAGACATTAG | TCCTCTGCCAGGATCACTTC |
| PPP3R1 | TGTAGACAAAACCATAATAAATGCAGA | GGATATCTAGGCCACCTACGAC |
| GAPDH | AGCTTGTCATCAACGGGAAG | TTTGATGTTAGTGGGGTCTCG |
CAMK1 = calcium/calmodulin-dependent protein kinase I; PLCE1 = phospholipase C, epsilon 1; PPP3CB (Calcineurin A beta) = protein phosphatase 3, catalytic subunit, beta isozyme; PPP3R1 (Calcineurin B) = protein phosphatase 3, regulatory subunit B, alpha; MEF2C = myocyte enhancer factor 2C.
Western Blotting
The protein extracts of human atrial tissues were examined by Western blot analysis. 20μg protein extracts were electrophoresed on 10% acrylamide SDS-PAGE gels and immunoblotted onto polyvinylidene difluoride membranes. The membranes were preblocked for 1 h in TBST (10 mM Tris–HCl pH 7.6, 150 mM NaCl, 0.1% Tween-20) containing 5% w/v nonfat dry milk and then incubated at 4°C overnight with anti-α-sarcomeric actin (Sigma Aldrich, Louis, MO, USA). The result of protein was normalized against GAPDH.
Histological Analysis
Atrial tissue sections were deparaffinized in xylene and rehydrated in decreasing concentrations of alcohol. Slides were then stained with hematoxylin/eosin. Tissue sections were observed under an Olympus BX51 microscope with the analysis including at least 100 randomly selected cells under 400 X magnification. All images of each specimen were captured using an Olympus DP70 camera. Atrial cardiomyocytes were analyzed (UTHSCSA, Image tool, Version 3.0).
Cell Culture and Mechanical Stretching of HL-1 Atrial Myocytes
HL-1 atrial myocytes were plated on silicone rubber culture dishes. HL-1 atrial myocytes were cultured for 24 hours in claycomb medium containing 10% FBS, penicillin and streptomycin. Thereafter, culture medium was changed to serum free claycomb medium and HL-1 atrial myocytes were stretched for an additional 8 hours in the same medium. HL-1 atrial myocytes received 15% uniaxial cyclic stretch at 1 Hz for 8 hours by NST-140 cell stretching system (NEPA GENE, Japan) in the cell stretched study groups. Stretched and control (non-stretched) experiments were carried out simultaneously with the same pool of cells in each experiment to match temperature, CO2 content, and pH of the medium for the stretched and control HL-1 atrial myocytes.
Immunofluorescence Staining
HL-1 atrial myocytes were fixed for 10 min with 4% paraformaldehyde, then exposed to 0.1% triton-X100, and stained with CytoPainter Phalloidin-iFluor 488 reagent (Abcam, Cambridge, USA), according to the manufacturer's protocol. Nuclei were stained with Hoechst 33258 (1:1000 dilution; Sigma, MO, USA). Four randomly chosen fields per section corresponding to at least fifty cells were examined at high magnification (400X). All images of each specimen were captured using a Leica DMI3000 microscope. Atrial cardiomyocytes were analyzed (UTHSCSA, Image tool, Version 3.0).
Statistical Analysis
Data are presented as mean ± SD (baseline characteristics) or SEM (gene and protein expressions). Categorical variables were compared using chi-square test or Fisher exact test as appropriate. Continuous variables among 3 groups were analyzed by the Kruskal-Wallis Test, and continuous variables between 2 groups were analyzed by the Mann-Whitney Test. Statistical analysis was performed using commercial statistical software (IBM SPSS Statistics 22). All P values were two-sided, and the level of statistical significance was set at 0.05.
Results
Baseline Characteristics of Patients Studied
Table 2 lists the clinical characteristics of the study patients with MR and patients with aortic valve disease. The two groups did not significantly differ in age, or heart failure status. The two groups also did not significantly differ in the preoperative left atrial ejection fraction, left ventricular size and left ventricular ejection fraction. However, the left atrial size was significantly larger in the MR patients than patients with aortic valve disease. Seventy-eight percent of MR patients and forty-two percent of patients with aortic valve disease received renin-angiotensin system blockers (P = 0.102).
Table 2. Baseline Clinical Characteristics of the Study Patients.
| MR (n = 14) | AVD (n = 7) | P value | |
|---|---|---|---|
| Age (years) | 58±9 | 60±11 | 0.550 |
| Male (%) | 6 (42.9%) | 6 (85.7%) | 0.159 |
| Body mass index (kg/m2) | 23.6±2.4 | 24.2±3.3 | 0.314 |
| Hypertension (%) | 7 (50.0%) | 4 (57.1%) | 1.000 |
| Diabetes mellitus (%) | 2 (14.3%) | 1 (14.3%) | 1.000 |
| Dyslipidemia (%) | 6 (42.9%) | 2 (28.6%) | 0.656 |
| Heart failure NYHA classification | 0.522 | ||
| Functional class I (%) | 2 (14.3%) | 1 (14.3%) | |
| Functional class II (%) | 6 (42.9%) | 3 (42.9%) | |
| Functional class III (%) | 6 (42.9%) | 2 (28.6%) | |
| Functional class IV (%) | 0 (0.0%) | 1 (14.3%) | |
| Tricuspid regurgitation (%) | 6 (42.9%) | 1 (14.3%) | 0.337 |
| Beta-blockers (%) | 4 (28.6%) | 0 (0.0%) | 0.255 |
| Calcium channel blockers (%) | 6 (42.9%) | 3 (42.9%) | 1.000 |
| Angiotensin converting enzyme inhibitors or angiotensin II receptor blockers (%) | 11 (78.6%) | 3 (42.9%) | 0.102 |
| Statins (%) | 1 (7.1%) | 0 (0.0%) | 1.000 |
| Creatinine (mg/dl) | 0.9±0.7 | 1.0±0.3 | 0.101 |
| White blood cell count (103/uL) | 6.3±1.6 | 5.6±1.8 | 0.331 |
| Left atrial diameter (mm) | 45.9±6.0 | 38.9±5.8 | 0.020 |
| Left atrial maximal volume (mL) | 88.1±44.1 | 42.5±25.6 | 0.037 |
| Left atrial ejection fraction (%) | 51.4±10.6 | 45.6±18.7 | 0.501 |
| Left ventricular end-diastolic diameter (mm) | 58.6±7.3 | 59.9±12.7 | 0.477 |
| Left ventricular ejection fraction (%) | 68.1±11.4 | 61.6±12.9 | 0.295 |
Data are presented as mean ± SD or number (percentage).
AVD = aortic valve disease; MR = mitral regurgitation; NYHA = New York Heart Association.
Identification and Enrichment Analysis of Differential Expression Genes between MR Patients and Normal Subjects
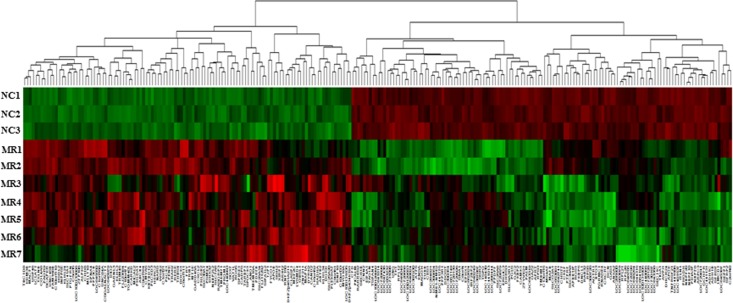
To determine the effect of MR on gene expression, we compared the expression profile in the left atrial free walls of the 7 MR patients to 3 normal subjects (76-year-old Caucasian female, 24-year-old Caucasian male and 27-year-old Caucasian male). A total of 244 differentially expressed genes were discovered by using genefilter R package [6] with the P value < 0.01 (t-test) and a fold-change cut-offs of > 1.5. A total of 112 genes were identified to be differentially up-regulated between MR patients and normal subjects (Table 3), and a total of 132 genes were identified to be differentially down-regulated between MR patients and normal subjects (Table 4), with the heat map graph being depicted in Fig 1. As with the unsupervised hierarchical clustering, the samples and genes were sorted corresponding to their respective groups.
Table 3. Selected Signature Upregulated Gene Expression in the Left Atria of Mitral Regurgitation vs. Normal Control.
| Symbol | Entrez ID | Gene Title | Gene Ontology | KEGG Pathway | Log2FC(MR/NC) |
|---|---|---|---|---|---|
| TMEM71 | 137835 | transmembrane protein 71 | CC: membrane | 1.879 | |
| DKFZp451A211 | 400169 | DKFZp451A211 protein | 1.787 | ||
| XIRP1 | 65904 | xin actin-binding repeat containing 1 | BP: negative regulation of cell proliferation, regulation of membrane potential, sarcomere organization, cardiac muscle cell development; CC: cell-cell adherens junction, fascia adherens; MF: actin binding, protein binding, poly(A) RNA binding | 1.689 | |
| PROM1 | 8842 | prominin 1 | CC: extracellular space, integral component of plasma membrane, cell surface, stereocilium, endoplasmic reticulum; MF: actinin binding, cadherin binding | 1.587 | |
| CXCL2 | 2920 | chemokine (C-X-C motif) ligand 2 | BP: immune response, inflammatory response, chemokine activity, positive regulation of leukocyte chemotaxis, chemokine-mediated signaling pathway, G-protein coupled receptor signaling pathway, positive regulation of cytosolic calcium ion concentration, regulation of cell proliferation, cell chemotaxis; CC: extracellular region, cytosol; MF: CXCR chemokine receptor binding, cytokine activity | TNF signaling pathway, Cytokine-cytokine receptor interaction, Chemokine signaling pathway, NF-kappa B signaling pathway, NOD-like receptor signaling pathway | 1.377 |
| PLCE1 | 51196 | phospholipase C, epsilon 1 | BP: activation of MAPK activity, calcium-mediated signaling, cell proliferation, cytoskeleton organization, diacylglycerol biosynthetic process, heart development, inositol phosphate metabolic process, phospholipase C-activating G-protein coupled receptor signaling pathway, positive regulation of cytosolic calcium ion concentration, protein kinase C-activating G-protein coupled receptor signaling pathway, Ras protein signal transduction, regulation of cell growth, regulation of G-protein coupled receptor protein signaling pathway, regulation of protein kinase activity; CC: cytoplasm, Golgi membrane, plasma membrane; MF: calcium ion binding, guanyl-nucleotide exchange factor activity, phosphatidylinositol phospholipase C activity, phospholipase C activity, protein binding, Ras GTPase binding, receptor signaling protein activity | Phosphatidylinositol signaling system, Ras signaling pathway, Inositol phosphate metabolism, Metabolic pathways, Rap1 signaling pathway, Calcium signaling pathway, cAMP signaling pathway | 1.333 |
| PLCXD3 | 345557 | phosphatidylinositol-specific phospholipase C, X domain containing 3 | BP: lipid metabolic process, signal transduction; MF: phosphoric diester hydrolase activity, signal transducer activity | 1.270 | |
| C10orf71 | 118461 | chromosome 10 open reading frame 71 | 1.200 | ||
| RGS5 | 8490 | regulator of G-protein signaling 5 | BP: positive regulation of GTPase activity, regulation of G-protein coupled receptor protein signaling pathway; CC: cytoplasm, plasma membrane; MF: GTPase activator activity | 1.168 | |
| C9orf61 | 9413 | chromosome 9 open reading frame 61 | 1.153 | ||
| CMYA5 | 202333 | cardiomyopathy associated 5 | BP: negative regulation of calcineurin-NFAT signaling cascade, negative regulation of protein phosphatase type 2B activity; CC: costamere, M band, perinuclear region of cytoplasm; MF: protein binding | 1.127 | |
| PLEKHA7 | 144100 | pleckstrin homology domain containing, family A member 7 | BP: epithelial cell-cell adhesion, zonula adherens maintenance; CC: centrosome, cytoplasm, zonula adherens; MF: delta-catenin binding | 1.119 | |
| GADD45A | 1647 | growth arrest and DNA-damage-inducible, alpha | BP: G2/M transition of mitotic cell cycle, activation of MAPKKK activity, negative regulation of protein kinase activity, cellular response to DNA damage stimulus, cell cycle arrest, centrosome cycle, signal transduction in response to DNA damage, positive regulation of apoptotic process, positive regulation of JNK cascade, positive regulation of p38MAPK cascade, regulation of cell cycle, positive regulation of reactive oxygen species metabolic process, negative regulation of protein kinase activity, cellular response to mechanical stimulus, DNA repair; CC: nucleus, cytoplasm; MF: core promoter binding, protein binding | FoxO signaling pathway, p53 signaling pathway, MAPK signaling pathway, Cell cycle | 1.117 |
| KLHL3 | 26249 | kelch-like 3 | BP: protein ubiquitination, protein ubiquitination involved in ubiquitin-dependent protein catabolic process, ion homeostasis, protein K48-linked ubiquitination; CC: cytosol, cytoskeleton, Cul3-RING ubiquitin ligase complex; MF: actin binding, catalytic activity | 1.094 | |
| DHX32 | 55760 | DEAH (Asp-Glu-Ala-His) box polypeptide 32 | BP: mRNA splicing, via spliceosome, RNA processing; CC: cytoplasm, spliceosomal complex, nucleus, mitochondrion, alpha DNA polymerase:primase complex; MF: poly(A) RNA binding, ATP-dependent RNA helicase activity, nucleotide binding, ATP binding | 1.076 | |
| LOC100133866 | 100133866 | 1.074 | |||
| PHLDB2 | 90102 | pleckstrin homology-like domain, family B, member 2 | CC: cytoplasm, plasma membrane, intermediate filament cytoskeleton; MF: protein binding | 1.064 | |
| HSDL2 | 84263 | hydroxysteroid dehydrogenase like 2 | CC: mitochondrion, peroxisome; MF: oxidoreductase activity, reduced coenzyme F420 dehydrogenase activity, NADPH:sulfur oxidoreductase activity, epoxyqueuosine reductase activity. malolactic enzyme activity, N-ethylmaleimide reductase activity | 1.056 | |
| DIO2 | 1734 | deiodinase, iodothyronine, type II | BP: oxidation-reduction process, thyroid hormone metabolic process, hormone biosynthetic process, response to hormone; CC: integral component of membrane; MF: thyroxine 5'-deiodinase activity, ubiquitin protein ligase binding | Thyroid hormone signaling pathway | 1.055 |
| MICAL2 | 9645 | microtubule associated monoxygenase, calponin and LIM domain containing 2 | BP: heart looping, cytoskeleton organization, positive regulation of transcription via serum response element binding, actin filament depolymerization, sulfur oxidation; CC: nucleus; MF: actin binding, oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen, NADPH:sulfur oxidoreductase activity. FAD binding, monooxygenase activity | 1.013 | |
| C15orf52 | 388115 | chromosome 15 open reading frame 52 | MF: poly(A) RNA binding | 1.013 | |
| MT1E | 4493 | metallothionein 1E | BP: negative regulation of growth, cellular response to cadmium ion, cellular response to zinc ion; CC: nucleus, cytoplasm; MF: zinc ion binding, metal ion binding | Mineral absorption | 1.008 |
| TBC1D8 | 11138 | TBC1 domain family, member 8 | BP: positive regulation of cell proliferation, blood circulation, positive regulation of Rab GTPase activity, regulation of cilium assembly; CC: membrane; MF: calcium ion binding, Rab GTPase activator activity | 1.001 | |
| CAND2 | 23066 | cullin-associated and neddylation-dissociated 2 (putative) | BP: SCF complex assembly, protein ubiquitination, positive regulation of transcription, DNA-templated; CC: nucleus; MF: protein binding, TBP-class protein binding | 0.996 | |
| THBS4 | 7060 | thrombospondin 4 | BP: positive regulation of endothelial cell proliferation, negative regulation of angiogenesis, regulation of tissue remodeling, response to endoplasmic reticulum stress, positive regulation of peptidyl-tyrosine phosphorylation, endothelial cell-cell adhesion, response to unfolded protein, tissue remodeling, positive regulation of cell division; CC: extracellular region, basement membrane, endoplasmic reticulum, sarcoplasmic reticulum, extracellular matrix, extracellular exosome; MF: integrin binding, calcium ion binding, protein binding, heparin binding, growth factor activity | ECM-receptor interaction, Phagosome, PI3K-Akt signaling pathway, Focal adhesion | 0.996 |
| APOB | 338 | apolipoprotein B | BP: retinoid metabolic process, receptor-mediated endocytosis, cholesterol metabolic process, positive regulation of lipid storage, low-density lipoprotein particle clearance; CC: extracellular region, cytoplasm, early endosome, endoplasmic reticulum lumen, Golgi apparatus, plasma membrane, actin cytoskeleton, clathrin-coated endocytic vesicle membrane; MF: protein binding, phospholipid binding, cholesterol transporter activity, lipase binding | Fat digestion and absorption, Vitamin digestion and absorption | 0.991 |
| ZFP106 | 64397 | zinc finger protein 106 homolog | BP: insulin receptor signaling pathway; CC: nucleolus, cytosol, membrane;MF: poly(A) RNA binding, opioid peptide activity, SH3 domain binding, metal ion binding | 0.969 | |
| RASGRP3 | 25780 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) | BP: Ras protein signal transduction, positive regulation of Ras GTPase activity, regulation of small GTPase mediated signal transduction, MAPK cascade; CC: cytoplasm, guanyl-nucleotide exchange factor complex, perinuclear region of cytoplasm; MF: Ras guanyl-nucleotide exchange factor activity, Ras GTPase binding, Rap GTPase activator activity, calcium ion binding, metal ion binding | Ras signaling pathway, MAPK signaling pathway, Rap1 signaling pathway | 0.966 |
| SLC25A34 | 284723 | solute carrier family 25, member 34 | BP: transport; CC: mitochondrion | 0.964 | |
| PHLDA1 | 22822 | pleckstrin homology-like domain, family A, member 1 | BP: apoptotic process, FasL biosynthetic process; CC: nucleolus, plasma membrane, cytoplasm; MF: protein binding | 0.961 | |
| SLC41A1 | 254428 | solute carrier family 41, member 1 | BP: cation transport, ion transport, transmembrane transport, CC: plasma membrane; MF: cation transmembrane transporter activity | 0.932 | |
| FLJ11292 | 55338 | 0.912 | |||
| RNF150 | 57484 | ring finger protein 150 | CC: membrane, integral component of membrane; MF: zinc ion binding, metal ion binding | 0.907 | |
| C6orf111 | 25957 | chromosome 6 open reading frame 111 | 0.904 | ||
| UNC84A | 23353 | unc-84 homolog A | CC: nuclear membrane, intracellular membrane-bounded organelle | 0.885 | |
| TRIM45 | 80263 | tripartite motif containing 45 | CC: nucleus, cytoplasm, intercellular bridge; MF: zinc ion binding, metal ion binding | 0.883 | |
| VAT1L | 57687 | vesicle amine transport protein 1 homolog (T. californica)-like | MF: oxidoreductase activity, zinc ion binding | 0.880 | |
| PTGFRN | 5738 | prostaglandin F2 receptor negative regulator | BP: lipid particle organization, negative regulation of translation; CC: endoplasmic reticulum, Golgi apparatus, cell surface, membrane; MF: protein binding | 0.871 | |
| CADPS | 8618 | Ca2+-dependent secretion activator | BP: transport, exocytosis, vesicle organization, synaptic vesicle priming, positive regulation of calcium ion-dependent exocytosis, catecholamine secretion, regulated secretory pathway; CC: membrane, cell junction, cytoplasmic vesicle, synapse; MF: calcium ion binding, protein binding, phosphatidylinositol-4,5-bisphosphate binding, lipid binding, protein kinase binding | 0.868 | |
| SIPA1L2 | 57568 | signal-induced proliferation-associated 1 like 2 | BP: positive regulation of GTPase activity, regulation of small GTPase mediated signal transduction; MF: GTPase activator activity | Rap1 signaling pathway | 0.865 |
| KLHL34 | 257240 | kelch-like 34 | CC: extracellular space | 0.860 | |
| ALPK2 | 115701 | alpha-kinase 2 | MF: ATP binding, protein serine/threonine kinase activity | 0.854 | |
| FAM13B | 51306 | family with sequence similarity 13, member B | BP: positive regulation of GTPase activity, regulation of mitochondrion degradation, small GTPase mediated signal transduction, regulation of small GTPase mediated signal transduction, activation of mitophagy in response to mitochondrial depolarization; CC: cytosol; MF: GTPase activator activity | 0.849 | |
| SOX7 | 83595 | SRY (sex determining region Y)-box 7 | BP: heart development, endoderm formation, negative regulation of cell proliferation, positive regulation of cysteine-type endopeptidase activity involved in apoptotic process, regulation of canonical Wnt signaling pathway, positive regulation of transcription, DNA-templated, negative regulation of transcription, DNA-templated, regulation of transcription from RNA polymerase II promoter; CC: nucleus, cytoplasm; MF: sequence-specific DNA binding transcription factor activity, transcription regulatory region DNA binding, RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity | 0.845 | |
| KLHL24 | 54800 | kelch-like 24 | BP: regulation of kainate selective glutamate receptor activity; CC: cytoplasm | 0.838 | |
| PYROXD2 | 84795 | pyridine nucleotide-disulphide oxidoreductase domain-containing protein 2 | BP: oxidation-reduction process; MF: oxidoreductase activity, N-ethylmaleimide reductase activity, reduced coenzyme F420 dehydrogenase activity, sulfur oxygenase reductase activity, malolactic enzyme activity, NADPH:sulfur oxidoreductase activity, epoxyqueuosine reductase activity, N-ethylmaleimide reductase activity | 0.821 | |
| INPP5E | 56623 | inositol polyphosphate-5-phosphatase | BP: phospholipid metabolic process, phosphatidylinositol biosynthetic process, phosphatidylinositol dephosphorylation, inositol phosphate dephosphorylation; CC: cytosol, axoneme, Golgi membrane, cytoskeleton; MF: inositol-polyphosphate 5-phosphatase activity, phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity, hydrolase activity | Inositol phosphate metabolism, Metabolic pathways, Phosphatidylinositol signaling system | 0.803 |
| TRAK2 | 66008 | trafficking protein, kinesin binding 2 | BP: regulation of transcription from RNA polymerase II promoter, protein O-linked glycosylation; CC: cytoplasm, mitochondrion, plasma membrane, nucleus, early endosome; MF: receptor binding, protein binding, GABA receptor binding, enzyme binding | Metabolic pathways, GABAergic synapse | 0.803 |
| TRIB1 | 10221 | 10221 | BP: positive regulation of proteasomal ubiquitin-dependent protein catabolic process, regulation of MAP kinase activity; CC: nucleus; MF: mitogen-activated protein kinase kinase binding | 0.800 | |
| LOC440993 | 440993 | 0.796 | |||
| SPOCK1 | 6695 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 | BP: negative regulation of cell-substrate adhesion, negative regulation of endopeptidase activity, signal transduction, neurogenesis; CC: extracellular space, cytoplasm, sarcoplasm, neuromuscular junction; MF: cysteine-type endopeptidase inhibitor activity, calcium ion binding, metalloendopeptidase inhibitor activity | 0.793 | |
| PENK | 5179 | proenkephalin | BP: neuropeptide signaling pathway, signal transduction; CC: extracellular region; MF: neuropeptide hormone activity, protein binding | 0.790 | |
| RERE | 473 | arginine-glutamic acid dipeptide (RE) repeats | BP: NLS-bearing protein import into nucleus, regulation of transcription, DNA-templated, chromatin remodeling; CC: nucleus, histone deacetylase complex; MF: chromatin binding, protein binding, poly-glutamine tract binding, sequence-specific DNA binding transcription factor activity, zinc ion binding | 0.769 | |
| MEF2C | 4208 | myocyte enhancer factor 2C | BP: negative regulation of transcription from RNA polymerase II promoter, positive regulation of transcription from RNA polymerase II promoter, MAPK cascade, positive regulation of gene expression, humoral immune response, positive regulation of myoblast differentiation, positive regulation of skeletal muscle tissue development, cellular response to lipopolysaccharide, cellular response to calcium ion, cellular response to transforming growth factor beta stimulus; CC: nucleus, cytoplasm, nuclear speck, protein complex; MF: RNA polymerase II regulatory region sequence-specific DNA binding, RNA polymerase II core promoter sequence-specific DNA binding transcription factor activity, DNA binding, miRNA binding | MAPK signaling pathway, cGMP-PKG signaling pathway | 0.767 |
| LOC645979 | 645979 | 0.765 | |||
| MAP1A | 4130 | microtubule-associated protein 1A | BP: activation of mitophagy in response to mitochondrial depolarization, microtubule cytoskeleton organization; CC: microtubule associated complex, cytosol, microtubule; MF: structural molecule activity, protein binding, microtubule binding | 0.760 | |
| FRY | 10129 | furry homolog | CC: spindle pole, cytoplasm, microtubule organizing center | 0.758 | |
| COL4A6 | 1288 | collagen, type IV, alpha 6 | BP: extracellular matrix disassembly, collagen catabolic process, extracellular matrix organization, cellular response to amino acid stimulus; CC: extracellular region, collagen type IV trimer, endoplasmic reticulum lumen, basement membrane; MF: extracellular matrix structural constituent, structural molecule activity | ECM-receptor interaction, PI3K-Akt signaling pathway, Focal adhesion | 0.757 |
| LCLAT1 | 253558 | lysocardiolipin acyltransferase 1 | BP: metabolic process, phospholipid metabolic process, phosphatidic acid biosynthetic process, triglyceride biosynthetic process, cardiolipin acyl-chain remodeling, glycerophospholipid biosynthetic process, CDP-diacylglycerol biosynthetic process; CC: endoplasmic reticulum, membrane; MF: transferase activity, transferring acyl groups, 1-acylglycerol-3-phosphate O-acyltransferase activity, sterol O-acyltransferase activity | Glycerophospholipid metabolism, Glycerolipid metabolism, Metabolic pathways | 0.755 |
| WSB2 | 55884 | WD repeat and SOCS box-containing 2 | BP: intracellular signal transduction, protein ubiquitination | 0.754 | |
| PSME4 | 23198 | proteasome (prosome, macropain) activator subunit 4 | BP: anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process, apoptotic process, cellular nitrogen compound metabolic process, cellular response to DNA damage stimulus, DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest, DNA repair, gene expression, mRNA metabolic process, multicellular organismal development, positive regulation of peptidase activity, proteasomal ubiquitin-independent protein catabolic process, protein polyubiquitination, regulation of cellular amino acid metabolic process; CC: cytosol, nucleus; MF: histone acetyl-lysine binding, peptidase activator activity | Proteasome | 0.752 |
| ABTB2 | 25841 | ankyrin repeat and BTB (POZ) domain containing 2 | BP: cellular response to toxic substance; CC: nucleus; MF: protein heterodimerization activity | 0.747 | |
| SLC7A6 | 9057 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 | BP: amino acid transport, blood coagulation, cellular amino acid metabolic process, ion transport, protein complex assembly, transmembrane transport; CC: plasma membrane. integral component of membrane; MF: amino acid transmembrane transporter activity, antiporter activity | 0.745 | |
| TOMM40L | 84134 | translocase of outer mitochondrial membrane 40 homolog (yeast)-like | BP: ion transport, protein transport, transmembrane transport; CC: mitochondrial outer membrane, pore complex, protein complex; MF: porin activity | 0.739 | |
| MYO18A | 399687 | myosin XVIIIA | BP: Golgi organization, cell migration, actomyosin structure organization, negative regulation of apoptotic process, Golgi vesicle budding, positive regulation of protein secretion, DNA metabolic process; CC: Golgi membrane, trans-Golgi network, actomyosin, myosin complex, endoplasmic reticulum-Golgi intermediate compartment; MF: protein binding, ATP binding, poly(A) RNA binding, actin filament binding, motor activity | 0.728 | |
| LOC100131835 | 100131835 | 0.718 | |||
| C1orf21 | 81563 | chromosome 1 open reading frame 21 | MF: protein binding | 0.714 | |
| KDM3B | 51780 | lysine (K)-specific demethylase 3B | BP: chromatin modification, transcription, DNA-templated; CC: nucleus; MF: dioxygenase activity, metal ion binding | 0.713 | |
| CLASP1 | 23332 | cytoplasmic linker associated protein 1 | BP: negative regulation of microtubule depolymerization; CC: cytoplasmic microtubule, kinetochore microtubule; MF: kinetochore binding, microtubule plus-end binding | 0.708 | |
| TMEM16A | 55107 | transmembrane protein 16A | BP: cation transport, chloride transport, ion transmembrane transport, regulation of membrane potential, phospholipase C-activating G-protein coupled receptor signaling pathway, regulation of anion transmembrane transport; CC: plasma membrane, extracellular vesicular exosome; MF: calcium activated cation channel activity, intracellular calcium activated chloride channel activity, protein binding, protein homodimerization activity, protein heterodimerization activity | 0.707 | |
| DGKD | 8527 | diacylglycerol kinase, delta 130kDa | BP: signal transduction, epidermal growth factor receptor signaling pathway, protein kinase C-activating G-protein coupled receptor signaling pathway, cell growth, diacylglycerol metabolic process, protein homooligomerization; CC: cytoplasm, plasma membrane, cytoplasmic membrane-bounded vesicle; MF: diacylglycerol kinase activity, protein binding, diacylglycerol binding, protein heterodimerization activity, protein homodimerization activity, NAD+ kinase activity, diacylglycerol kinase activity, ATP binding | Glycerolipid metabolism, Glycerophospholipid metabolism, Metabolic pathways, Phosphatidylinositol signaling system | 0.693 |
| HERC2 | 8924 | HECT and RLD domain containing E3 ubiquitin protein ligase 2 | BP: DNA repair, intracellular protein transport, protein ubiquitination, regulation of GTPase activity; CC: cytoplasm, mitochondrial inner membrane, nucleus; MF: guanyl-nucleotide exchange factor activity, heme binding, protein binding, SUMO binding, ubiquitin protein ligase binding, ubiquitin-protein ligase activity | Ubiquitin mediated proteolysis | 0.687 |
| DKFZp434K191 | 29797 | 0.686 | |||
| KBTBD12 | 166348 | kelch repeat and BTB (POZ) domain containing 12 | 0.683 | ||
| DOCK1 | 1793 | dedicator of cytokinesis 1 | BP: cytoskeleton organization, small GTPase mediated signal transduction, cell migration, positive regulation of GTPase activity; MF: guanyl-nucleotide exchange factor activity, GTPase activator activity; CC: intracellular | Focal adhesion, Regulation of actin cytoskeleton | 0.681 |
| C1orf168 | 199920 | chromosome 1 open reading frame 168 | 0.674 | ||
| RNF10 | 9921 | ring finger protein 10 | BP: negative regulation of Schwann cell proliferation, positive regulation of myelination, positive regulation of transcription from RNA polymerase II promoter, positive regulation of transcription, DNA-templated; CC: nucleus, cytoplasm; MF: transcription regulatory region DNA binding, zinc ion binding, protein binding | 0.671 | |
| CHD6 | 84181 | chromodomain helicase DNA binding protein 6 | BP: positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress, metabolic process, transcription, DNA-templated; CC: nucleoplasm, DNA-directed RNA polymerase II, core complex; MF: transcription cofactor binding, DNA-dependent ATPase activity, DNA binding, chromatin binding, ATP binding, ATP-dependent helicase activity | 0.669 | |
| USP46 | 64854 | ubiquitin specific peptidase 46 | BP: ubiquitin-dependent protein catabolic process, protein deubiquitination; MF: ubiquitin thiolesterase activity, protein binding, ubiquitin-specific protease activity | 0.668 | |
| C10orf110 | 55853 | chromosome 10 open reading frame 110 | 0.667 | ||
| UNC45B | 146862 | unc-45 homolog B | BP: chaperone-mediated protein folding, cardiac muscle tissue development, cell differentiation, myofibril assembly; CC: cytosol, Z disc, A band; MF: Hsp90 protein binding | 0.662 | |
| KPNA4 | 3840 | karyopherin alpha 4 (importin alpha 3) | BP: protein import into nucleus, response to hydrogen peroxide, NLS-bearing protein import into nucleus, cytokine-mediated signaling pathway, protein transport; CC: nucleus, cytoplasm, extracellular vesicular exosome; MF: protein transporter activity, protein binding | 0.655 | |
| SEMA4D | 349236 | Semaphorin-4D | BP: negative regulation of transcription from RNA polymerase II promoter, positive regulation of protein phosphorylation, negative regulation of cell adhesion, regulation of cell shape, negative regulation of alkaline phosphatase activity, positive regulation of phosphatidylinositol 3-kinase signaling, positive regulation of cell migration, positive regulation of GTPase activity, positive regulation of peptidyl-tyrosine phosphorylation, negative regulation of peptidyl-tyrosine phosphorylation; CC: extracellular space, plasma membrane; MF: receptor activity, transmembrane signaling receptor activity, receptor binding | 0.654 | |
| GARNL3 | 84253 | GTPase activating Rap/RanGAP domain-like 3 | BP: positive regulation of GTPase activity, regulation of small GTPase mediated signal transduction; MF: GTPase activator activity | 0.650 | |
| TJP1 | 7082 | tight junction protein 1 | BP: tight junction assembly, apoptotic process, cellular component disassembly involved in execution phase of apoptosis; CC: cytoplasm, plasma membrane, cell junction, tight junction, cytoplasmic vesicle, intercalated disc; MF: protein binding, protein domain specific binding | Adherens junction, Tight junction, Gap junction | 0.645 |
| LOC645550 | 645550 | 0.644 | |||
| MACF1 | 23499 | microtubule-actin crosslinking factor 1 | BP: cell cycle arrest, metabolic process, Wnt signaling pathway, posttranslational protein targeting to membrane, establishment or maintenance of cell polarity, Golgi to plasma membrane protein transport; CC: cytoskeleton, Golgi apparatus; MF: actin binding, calcium ion binding, microtubule binding, ATPase activity, poly(A) RNA binding | 0.642 | |
| ANO1 | 55107 | anoctamin 1, calcium activated chloride channel | BP: regulation of membrane potential, cation transmembrane transport, chloride transmembrane transport; CC: external side of plasma membrane, integral component of membrane, extracellular exosome; MF: calcium activated cation channel activity, intracellular calcium activated chloride channel activity, protein homodimerization activity, protein heterodimerization activity | 0.638 | |
| ZNF211 | 10520 | zinc finger protein 211 | BP: regulation of transcription, DNA-templated; CC: nucleus; MF: nucleic acid binding, metal ion binding, DNA binding | 0.637 | |
| PAQR9 | 344838 | progestin and adipoQ receptor family member IX | CC: integral component of membrane; MF: receptor activity | 0.637 | |
| FYCO1 | 79443 | FYVE and coiled-coil domain containing 1 | BP: transport; CC: integral component of membrane | 0.636 | |
| ARIH2 | 10425 | ariadne RBR E3 ubiquitin protein ligase 2 | BP: protein polyubiquitination, protein ubiquitination involved in ubiquitin-dependent protein catabolic process, developmental cell growth, protein K63-linked ubiquitination, protein K48-linked ubiquitination; CC: nucleus, cytoplasm; MF: ubiquitin-protein transferase activity, protein binding, zinc ion binding, nucleic acid binding, ligase activity | 0.635 | |
| SLC30A1 | 7779 | solute carrier family 30 (zinc transporter), member 1 | BP: zinc II ion transport, cellular calcium ion homeostasis, negative regulation of calcium ion import, cellular zinc ion homeostasis, transmembrane transport, cadmium ion transmembrane transport; CC: cytoplasm, endoplasmic reticulum, Golgi apparatus, T-tubule, nuclear membrane, plasma membrane; MF: protein binding, calcium channel inhibitor activity, cation transmembrane transporter activity | Mineral absorption | 0.635 |
| SSH2 | 85464 | slingshot homolog 2 protein phosphatase | BP: actin cytoskeleton organization, protein dephosphorylation, regulation of actin polymerization or depolymerization, regulation of axonogenesis; CC: cytoplasm, cytoskeleton; MF: DNA binding, protein tyrosine phosphatase activity, protein tyrosine/serine/threonine phosphatase activity, actin binding, phosphoprotein phosphatase activity | Regulation of actin cytoskeleton | 0.628 |
| HADHA | 3030 | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit | BP: fatty acid beta-oxidation, phospholipid metabolic process, cardiolipin acyl-chain remodeling, glycerophospholipid biosynthetic process, fatty acid metabolic process, oxidation-reduction process; CC: mitochondrion, mitochondrial inner membrane, mitochondrial fatty acid beta-oxidation multienzyme complex; MF: 3-hydroxyacyl-CoA dehydrogenase activity, acetyl-CoA C-acetyltransferase activity, enoyl-CoA hydratase activity, protein binding, long-chain-3-hydroxyacyl-CoA dehydrogenase activity, coenzyme binding, NAD binding | Fatty acid metabolism, Metabolic pathways, Fatty acid elongation, Fatty acid degradation, Biosynthesis of unsaturated fatty acids, Valine, leucine and isoleucine degradation, Lysine degradation, Tryptophan metabolism, beta-Alanine metabolism, Propanoate metabolism, Butanoate metabolism | 0.624 |
| CARKD | 55739 | carbohydrate kinase domain containing | BP: nicotinamide nucleotide metabolic process; CC: mitochondrion; MF: ATP binding, ATP-dependent NAD(P)H-hydrate dehydratase activity, lyase activity | 0.623 | |
| DYRK2 | 8445 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | BP: intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator, cellular response to DNA damage stimulus, protein phosphorylation, peptidyl-tyrosine phosphorylation, positive regulation of glycogen biosynthetic process, negative regulation of NFAT protein import into nucleus; CC: nucleus, cytoplasm, ubiquitin ligase complex, ribonucleoprotein complex; MF: magnesium ion binding, protein serine/threonine kinase activity, protein tyrosine kinase activity, ATP binding, ubiquitin binding | 0.622 | |
| SPIRE1 | 56907 | spire homolog 1 | BP: Golgi vesicle transport, actin cytoskeleton organization, protein transport, actin nucleation; CC: Golgi apparatus, cytoplasmic vesicle membrane, cytoskeleton, Golgi apparatus, plasma membrane; MF: actin binding | 0.621 | |
| FAM168B | 130074 | family with sequence similarity 168, member B | CC: extracellular vesicular exosome, plasma membrane, perinuclear region of cytoplasm, plasma membrane | 0.611 | |
| PDE7B | 27115 | phosphodiesterase 7B | BP: cAMP-mediated signaling, signal transduction; CC: cytosol; MF: 3',5'-cyclic-AMP phosphodiesterase activity, metal ion binding, phosphoric diester hydrolase activity | Purine metabolism | 0.610 |
| CSGALNACT1 | 55790 | chondroitin sulfate N-acetylgalactosaminyltransferase 1 | BP: UDP-N-acetylgalactosamine metabolic process, extracellular matrix organization, UDP-glucuronate metabolic process, cartilage development; CC: Golgi cisterna membrane; MF: peptidoglycan glycosyltransferase activity, glucuronosyltransferase activity, glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity | Glycosaminoglycan biosynthesis—chondroitin sulfate/dermatan sulfate, Metabolic pathways | 0.609 |
| MLLT10 | 8028 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | BP: positive regulation of transcription from RNA polymerase II promoter, transcription, DNA-templated, canonical Wnt signaling pathway; CC: nucleus, cytoplasm; MF: zinc ion binding, DNA binding, sequence-specific DNA binding transcription factor activity | 0.608 | |
| N4BP2L2 | 10443 | NEDD4 binding protein 2-like 2 | BP: negative regulation of transcription from RNA polymerase II promoter; CC: nucleus, transcriptional repressor complex, extracellular vesicular exosome, cytoplasm; MF: RNA polymerase II transcription corepressor activity, protein binding, enzyme binding | 0.608 | |
| NARS | 4677 | asparaginyl-tRNA synthetase | BP:asparaginyl-tRNA aminoacylation, tRNA aminoacylation for protein translation, translation; CC: cytoplasm, mitochondrion, extracellular exosome; MF: nucleic acid binding, asparagine-tRNA ligase activity, aminoacyl-tRNA ligase activity, ATP binding | Aminoacyl-tRNA biosynthesis | 0.606 |
| LYSMD4 | 145748 | LysM, putative peptidoglycan-binding, domain containing 4 | CC: integral component of membrane, membrane | 0.606 | |
| TXLNB | 167838 | taxilin beta | BP: positive regulation of neuron projection development; CC: cytoplasm; MF: syntaxin binding | 0.604 | |
| TARBP1 | 6894 | TAR (HIV-1) RNA binding protein 1 | BP: regulation of transcription from RNA polymerase II promoter, RNA methylation, RNA processing; CC: nucleus; MF: RNA binding, RNA methyltransferase activity | 0.601 | |
| FLJ23584 | 79640 | 0.596 | |||
| SCAPER | 49855 | S-phase cyclin A-associated protein in the ER | CC: nucleus, cytoplasm, endoplasmic reticulum; MF: metal ion binding, nucleic acid binding | 0.594 | |
| OBSL1 | 23363 | obscurin-like 1 | BP: microtubule cytoskeleton organization, Golgi organization, regulation of mitotic nuclear division, protein localization to Golgi apparatus, cardiac myofibril assembly; CC: Golgi apparatus, centrosome, cytoplasm, 3M complex; MF: cytoskeletal adaptor activity | 0.585 | |
| SLC25A20 | 788 | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 | BP: cellular lipid metabolic process, transport, small molecule metabolic process; CC: mitochondrial inner membrane, mitochondrion | 0.584 | |
| PPP3CB | 5532 | protein phosphatase 3, catalytic subunit, beta isozyme | CC: calcineurin complex, nucleus; MF: calcium channel inhibitor activity, calcium ion binding, calmodulin binding, enzyme binding, protein phosphatase 2B binding, phosphoprotein phosphatase activity, calmodulin-dependent protein phosphatase activity | MAPK signaling pathway, Calcium signaling pathway, cGMP-PKG signaling pathway, Apoptosis, Wnt signaling pathway, VEGF signaling pathway | 0.581 |
| ABCB4 | 5244 | ATP-binding cassette, sub-family B (MDR/TAP), member 4 | BP: transmembrane transport; CC: integral component of membrane, extracellular vesicular exosome, mitochondrion; MF: ATPase activity, coupled to transmembrane movement of substances, ATP binding, nucleotide binding, transporter activity | ABC transporters | 0.580 |
BP: biological process; CC: cell component; MF: molecular function.
Table 4. Selected Signature Downregulated Gene Expressions in the Left Atria of Mitral Regurgitation vs. Normal Control.
| Symbol | Entrez ID | Gene Title | Gene Ontology | KEGG Pathway | Log2FC(MR/NC) |
|---|---|---|---|---|---|
| ITLN1 | 55600 | intelectin 1 (galactofuranose binding) | BP: positive regulation of protein phosphorylation, positive regulation of glucose import; CC: anchored component of membrane, receptor complex, membrane raft, extracellular exosome; MF: carbohydrate binding | -5.119 | |
| NBPF20 | 400818 | neuroblastoma breakpoint family, member 20, transcript variant 4 | CC: cytoplasm | -2.848 | |
| TKT | 7086 | transketolase (Wernicke-Korsakoff syndrome) | BP: metabolic process; MF: catalytic activity | Carbon metabolism, Biosynthesis of amino acids, Metabolic pathways, Pentose phosphate pathway | -2.402 |
| SFRP2 | 6423 | secreted frizzled-related protein 2 | BP: patterning of blood vessels, cardiac left ventricle morphogenesis, cell-cell signaling, response to nutrient, positive regulation of cell proliferation, negative regulation of gene expression, negative regulation of cardiac muscle cell apoptotic process, positive regulation of endopeptidase activity, negative regulation of Wnt signaling pathway, collagen fibril organization, positive regulation of cell growth, negative regulation of cell growth, negative regulation of cell migration, negative regulation of BMP signaling pathway, cellular response to extracellular stimulus, positive regulation of peptidyl-serine phosphorylation, positive regulation of cell adhesion mediated by integrin, positive regulation of catenin import into nucleus, non-canonical Wnt signaling pathway, positive regulation of apoptotic process, negative regulation of JUN kinase activity, negative regulation of transcription, DNA-templated, canonical Wnt signaling pathway, negative regulation of extrinsic apoptotic signaling pathway via death domain receptors, negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage, negative regulation of planar cell polarity pathway involved in axis elongation; CC: extracellular space, extracellular matrix; MF: fibronectin binding, integrin binding, metalloenzyme activator activity, Wnt-protein binding, receptor agonist activity, endopeptidase activator activity | Wnt signaling pathway | -2.290 |
| ARPC3 | 10094 | actin related protein 2/3 complex, subunit 3 | BP: movement of cell or subcellular component, Arp2/3 complex-mediated actin nucleation, Fc-gamma receptor signaling pathway involved in phagocytosis, ephrin receptor signaling pathway, actin filament organization; CC: cytosol, Arp2/3 protein complex, actin cytoskeleton, extracellular vesicular exosome, cell projection, lamellipodium; MF: structural constituent of cytoskeleton, protein binding, actin filament binding | Regulation of actin cytoskeleton, Fc gamma R-mediated phagocytosis | -2.124 |
| SLCO2A1 | 6578 | solute carrier organic anion transporter family, member 2A1 | BP: lipid transport, prostaglandin transport, sodium-independent organic anion transport, transmembrane transport; CC: plasma membrane; MF: lipid transporter activity, prostaglandin transmembrane transporter activity | -2.115 | |
| LOC729841 | 729841 | -1.872 | |||
| NBPF10 | 440673 | neuroblastoma breakpoint family, member 10, transcript variant 5 | CC: cytoplasm; MF: poly(A) RNA binding | -1.783 | |
| RGS1 | 5996 | regulator of G-protein signaling 1 | BP: immune response, signal transduction, adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway, termination of G-protein coupled receptor signaling pathway, positive regulation of GTPase activity; CC: cytoplasm, plasma membrane; MF: GTPase activator activity, calmodulin binding | -1.764 | |
| ADH1B | 125 | alcohol dehydrogenase IB (class I), beta polypeptide | BP: oxidation-reduction process; MF: zinc ion binding, oxidoreductase activity | Metabolic pathways, Glycolysis/Gluconeogenesis, Fatty acid degradation, Tyrosine metabolism | -1.727 |
| APOE | 348 | apolipoprotein E | BP: cholesterol biosynthetic process, cholesterol catabolic process, triglyceride catabolic process, cholesterol transport, lipoprotein catabolic process, neuron projection regeneration; CC: extracellular region; MF: cholesterol binding, cholesterol transporter activity | -1.716 | |
| RPS3 | 6188 | ribosomal protein S3 | BP: DNA catabolic process, endonucleolytic, cytoplasmic translation, DNA repair; CC: ribosome, nucleus, cytosolic small ribosomal subunit; MF: structural constituent of ribosome, RNA binding, damaged DNA binding, oxidized purine nucleobase lesion DNA N-glycosylase activity, protein kinase A binding | Ribosome | -1.604 |
| LOC388654 | 388654 | -1.477 | |||
| LOC651149 | 651149 | -1.434 | |||
| LOC390354 | 390354 | -1.422 | |||
| LOC729926 | 729926 | -1.399 | |||
| C8orf4 | 56892 | chromosome 8 open reading frame 4 | BP: apoptotic process | -1.397 | |
| RARRES2 | 5919 | retinoic acid receptor responder (tazarotene induced) 2 | BP: inflammatory response, regulation of lipid catabolic process; MF: receptor binding | -1.310 | |
| TMBIM1 | 64114 | transmembrane BAX inhibitor motif containing 1 | BP: negative regulation of extrinsic apoptotic signaling pathway via death domain receptors, negative regulation of Fas signaling pathway, negative regulation of establishment of protein localization to plasma membrane; CC: integral component of membrane, Golgi apparatus, lysosomal membrane, extracellular vesicular exosome; MF: death receptor binding | -1.295 | |
| NBPF8 | 728841 | neuroblastoma breakpoint family, member 8 | CC: cytoplasm | -1.281 | |
| PPP3R1 | 5534 | protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform, Calcineurin subunit B type 1 | BP: stimulatory C-type lectin receptor signaling pathway, apoptotic process, dephosphorylation, calcineurin-NFAT signaling cascade, Fc-epsilon receptor signaling pathway, innate immune response, positive regulation of transcription from RNA polymerase II promoter, positive regulation of NFAT protein import into nucleus, intrinsic apoptotic signaling pathway, positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway; CC: sarcolemma, nucleoplasm, cytosol, calcineurin complex; MF: calcium ion binding, calcium-dependent protein serine/threonine phosphatase activity, calcium ion binding, protein binding, calmodulin binding, protein domain specific binding | MAPK signaling pathway, Calcium signaling pathway, cGMP-PKG signaling pathway, Apoptosis, Wnt signaling pathway, VEGF signaling pathway, Glucagon signaling pathway | -1.267 |
| DDOST | 1650 | dolichyl-diphosphooligosaccharide-protein glycosyltransferase | BP: protein N-linked glycosylation via asparagine, cellular protein metabolic process, gene expression, post-translational protein modification, protein glycosylation, translation; CC: endoplasmic reticulum membrane, intracellular membrane-bounded organelle; MF: dolichyl-diphosphooligosaccharide-protein glycotransferase activity, oligosaccharyl transferase activity, protein binding | Metabolic pathways, N-Glycan biosynthesis, Protein processing in endoplasmic reticulum | -1.264 |
| ZFYVE21 | 79038 | zinc finger, FYVE domain containing 21 | CC: endosome, focal adhesion, cytoplasmic membrane-bounded vesicle; MF: protein binding, metal ion binding | -1.255 | |
| PDIA3P | 171423 | protein disulfide isomerase family A, member 3 pseudogene | -1.245 | ||
| LOC441131 | 441131 | -1.237 | |||
| LOC654194 | 654194 | -1.234 | |||
| SLC39A8 | 64116 | solute carrier family 39 (zinc transporter), member 8 | BP: transmembrane transport, zinc ion transport; CC: integral component of membrane; MF: metal ion transmembrane transporter activity | -1.230 | |
| TAGLN | 6876 | transgelin | BP: muscle organ development, epithelial cell differentiation; CC: cytoplasm; MF: actin filament binding, protein binding | -1.223 | |
| FKBP5 | 2289 | FK506 binding protein 5 | BP: protein peptidyl-prolyl isomerization, protein folding, chaperone-mediated protein folding; CC: nucleoplasm, cytoplasm, endoplasmic reticulum membrane, extracellular exosome; MF: peptidyl-prolyl cis-trans isomerase activity, protein binding, FK506 binding, heat shock protein binding | Estrogen signaling pathway | -1.213 |
| LOC648024 | 648024 | -1.193 | |||
| LOC387867 | 387867 | -1.182 | |||
| LOC100131905 | 100131905 | -1.139 | |||
| PHGDH | 26227 | phosphoglycerate dehydrogenase | BP: cell cycle process, cellular amino acid biosynthetic process, cellular nitrogen compound metabolic process, glutamine metabolic process, regulation of gene expression, small molecule metabolic process, taurine metabolic process, threonine metabolic process, L-serine biosynthetic process; CC: cytosol; MF: electron carrier activity, NAD binding, phosphoglycerate dehydrogenase activity | Biosynthesis of amino acids, Carbon metabolism, Glycine, serine and threonine metabolism, Metabolic pathways | -1.132 |
| LOC728139 | 728139 | -1.126 | |||
| LOC728698 | 728698 | -1.107 | |||
| LOC284821 | 284821 | -1.106 | |||
| EPHX1 | 2052 | epoxide hydrolase 1, microsomal | BP: cellular aromatic compound metabolic process; CC: endoplasmic reticulum membrane; MF: cis-stilbene-oxide hydrolase activity, epoxide hydrolase activity | -1.081 | |
| PICALM | 8301 | phosphatidylinositol binding clathrin assembly protein | BP: clathrin coat assembly; CC: clathrin coat, intracellular membrane-bounded organelle; MF: 1-phosphatidylinositol binding, clathrin binding, phospholipid binding | -1.081 | |
| CFD | 1675 | complement factor D | BP: proteolysis, complement activation, blood coagulation, platelet degranulation, platelet activation; CC: extracellular region, platelet alpha granule lumen extracellular exosome; MF: serine-type endopeptidase activity | Complement and coagulation cascades | -1.072 |
| GAS1 | 2619 | growth arrest-specific 1 | BP: regulation of smoothened signaling pathway, negative regulation of protein processing, cellular response to vascular endothelial growth factor stimulus, regulation of apoptotic process, cell fate commitment, negative regulation of mitotic cell cycle, developmental growth, regulation of ER to Golgi vesicle-mediated transport, positive regulation of mesenchymal cell proliferation, cell cycle arrest, negative regulation of cell growth, positive regulation of epithelial cell proliferation, negative regulation of epithelial cell proliferation, negative regulation of extrinsic apoptotic signaling pathway in absence of ligand; CC: plasma membrane, integral component of membrane; MF: protein binding | Hedgehog signaling pathway | -1.067 |
| FKBP2 | 2286 | FK506 binding protein 2 | BP: protein folding, peptidyl-proline modification;CC: endoplasmic reticulum, membrane; MF: peptidyl-prolyl cis-trans isomerase activity, FK506 binding, protein binding | -1.058 | |
| RPL21 | 6144 | ribosomal protein L21 | BP: translation;CC: nucleolus, cytoplasm, ribosome; MF: structural constituent of ribosome, poly(A) RNA binding | Ribosome | -1.044 |
| RNASE4 | 6038 | ribonuclease, RNase A family, 4 | BP: mRNA cleavage, RNA phosphodiester bond hydrolysis, endonucleolytic; CC: extracellular region, extracellular exosome; MF: nucleic acid binding, ribonuclease A activity | -1.039 | |
| OAZ2 | 4947 | ornithine decarboxylase antizyme 2 | BP: cellular nitrogen compound metabolic process, negative regulation of catalytic activity, polyamine metabolic process, regulation of cellular amino acid metabolic process, small molecule metabolic process; CC: cytosol, nucleus; MF: ornithine decarboxylase inhibitor activity | -1.025 | |
| MAGOH | 4116 | mago-nashi homolog, proliferation-associated | BP: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay, regulation of alternative mRNA splicing, via spliceosome, mRNA splicing, via spliceosome, transcription from RNA polymerase II promoter, termination of RNA polymerase II transcription, mRNA export from nucleus, regulation of translation, RNA splicing, gene expression, mRNA 3'-end processing; CC: nucleus, nucleoplasm, cytosol, nuclear speck, exon-exon junction complex, catalytic step 2 spliceosome; MF: protein binding, poly(A) RNA binding | RNA transport, mRNA surveillance pathway, Spliceosome | -1.019 |
| LOC648249 | 648249 | -1.004 | |||
| PTPRF | 5792 | protein tyrosine phosphatase, receptor type, F | BP: peptidyl-tyrosine dephosphorylation, cell adhesion, transmembrane receptor protein tyrosine phosphatase signaling pathway, cell migration, negative regulation of receptor binding; CC: integral component of plasma membrane, extracellular exosome; MF: protein tyrosine phosphatase activity, transmembrane receptor protein tyrosine phosphatase activity, protein complex binding | Cell adhesion molecules, Adherens junction, Insulin signaling pathway | -0.997 |
| LOC441775 | 441775 | -0.991 | |||
| LOC440055 | 440055 | -0.990 | |||
| LOC100129553 | 100129553 | -0.978 | |||
| RAB32 | 10981 | RAB32, member RAS oncogene family | BP: intracellular protein transport, metabolic process, Rab protein signal transduction, endosome to melanosome transport, phagosome maturation; CC: mitochondrion, early endosome, trans-Golgi network, membrane, phagocytic vesicle membrane, melanosome; MF: GTPase activity, protein binding, GTP binding, GTP-dependent protein binding, AP-1 adaptor complex binding, AP-3 adaptor complex binding, BLOC-2 complex binding | -0.965 | |
| CHST7 | 56548 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 | BP: carbohydrate metabolic process, polysaccharide metabolic process, N-acetylglucosamine metabolic process, sulfur compound metabolic process, glycosaminoglycan metabolic process, chondroitin sulfate metabolic process; CC: Golgi membrane, integral component of membrane; MF: N-acetylglucosamine 6-O-sulfotransferase activity, chondroitin 6-sulfotransferase activity | Glycosaminoglycan biosynthesis—chondroitin sulfate / dermatan sulfate | -0.961 |
| DHCR24 | 1718 | 24-dehydrocholesterol reductase | BP: cholesterol biosynthetic process, apoptotic process, negative regulation of apoptotic process, negative regulation of cysteine-type endopeptidase activity involved in apoptotic process, response to oxidative stress, oxidation-reduction process, cell cycle arrest, Ras protein signal transduction, protein localization, negative regulation of cell proliferation, plasminogen activation, amyloid precursor protein catabolic process; CC: Golgi membrane, nucleus, cytoplasm, endoplasmic reticulum, cytoskeleton; MF: delta24(24–1) sterol reductase activity, UDP-N-acetylmuramate dehydrogenase activity, oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor, enzyme binding, peptide antigen binding, flavin adenine dinucleotide binding | Steroid biosynthesis, Metabolic pathways | -0.952 |
| GCHFR | 2644 | GTP cyclohydrolase I feedback regulator | BP: negative regulation of biosynthetic process, negative regulation of GTP cyclohydrolase I activity; CC: cytoplasm, nucleus, protein complex; MF: amino acid binding, enzyme inhibitor activity, GTP cyclohydrolase binding, GTP-dependent protein binding, hydrolase activity | -0.939 | |
| LOC643319 | 643319 | -0.939 | |||
| DCN | 1634 | decorin | MF: collagen binding | TGF-beta signaling pathway | -0.936 |
| LOC653079 | 653079 | -0.927 | |||
| DNCL1 | 8655 | dynein, cytoplasmic, light polypeptide 1 | BP: transcription, DNA-templated, regulation of transcription, DNA-templated, transport, microtubule-based process, regulation of catalytic activity, G2/M transition of mitotic cell cycle, mitotic cell cycle, apoptotic process, organelle organization, actin cytoskeleton organization, negative regulation of phosphorylation, positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway, intrinsic apoptotic signaling pathway; CC: mitochondrion, microtubule, nucleus, cytoplasm, cytoplasmic dynein complex, extracellular exosome, mitotic spindle, COP9 signalosome; MF: motor activity, enzyme binding, nitric-oxide synthase regulator activity, protein homodimerization activity, protein binding | -0.922 | |
| SEPP1 | 6414 | selenoprotein P, plasma, 1 | BP: selenium compound metabolic process, growth, response to oxidative stress; CC: extracellular space, extracellular exosome; MF: selenium binding | -0.917 | |
| MAFB | 9935 | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B | BP: positive regulation of transcription from RNA polymerase II promoter, transcription, DNA-templated; CC: nucleus, transcription factor complex; MF: sequence-specific DNA binding transcription factor activity, sequence-specific DNA binding, transcription factor binding, DNA binding | -0.910 | |
| LOC441506 | 441506 | -0.905 | |||
| NME1-NME2 | 654364 | NME1-NME2 readthrough | BP: nucleoside diphosphate phosphorylation, GTP biosynthetic process, negative regulation of gene expression, regulation of apoptotic process; CC: centrosome, cytosol, mitochondrion; MF: RNA polymerase II regulatory region sequence-specific DNA binding, single-stranded DNA binding, nucleoside diphosphate kinase activity, ATP binding, protein kinase binding | Metabolic pathways, Purine metabolism, Pyrimidine metabolism | -0.905 |
| CXCR7 | 57007 | chemokine (C-X-C motif) receptor 7 | BP: angiogenesis, vasculogenesis, cell adhesion, G-protein coupled receptor signaling pathway, receptor internalization, chemokine-mediated signaling pathway, positive regulation of ERK1 and ERK2 cascade, negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage, chemotaxis; CC: endosome, plasma membrane, perinuclear region of cytoplasm; MF: scavenger receptor activity, coreceptor activity, C-X-C chemokine receptor activity, signal transducer activity | -0.903 | |
| LOC653881 | 653881 | -0.887 | |||
| SIVA1 | 10572 | SIVA1, apoptosis-inducing factor | BP: negative regulation of NF-kappaB transcription factor activity, extrinsic apoptotic signaling pathway, intrinsic apoptotic signaling pathway, positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway; CC: cytoplasm, nucleoplasm, mitochondrion; MF: CD27 receptor binding, zinc ion binding, tumor necrosis factor receptor binding | -0.863 | |
| LOC286444 | 286444 | -0.860 | |||
| LOC100128892 | 100128892 | -0.859 | |||
| LOC728658 | 728658 | -0.859 | |||
| LOC401537 | 401537 | -0.837 | |||
| C2orf40 | 84417 | chromosome 2 open reading frame 40 | BP: negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation, G1 to G0 transition, cellular senescence; CC: extracellular space, transport vesicle | -0.820 | |
| LOC649049 | 649049 | -0.812 | |||
| C13orf15 | 28984 | chromosome 13 open reading frame 15 | BP: negative regulation of exit from mitosis, negative regulation of endothelial cell proliferation, positive regulation of extracellular matrix constituent secretion, complement activation, positive regulation of epithelial to mesenchymal transition, negative regulation of angiogenesis, positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle, positive regulation of collagen biosynthetic process, positive regulation of mitotic nuclear division, positive regulation of transcription from RNA polymerase II promoter, negative regulation of cytokine secretion, positive regulation of cytokine secretion, positive regulation of sequence-specific DNA binding transcription factor activity, positive regulation of stress fiber assembly, positive regulation of cell cycle arrest, mitotic cell cycle arrest, negative regulation of mitotic cell cycle phase transition; CC: nucleus, cytoplasm, centrosome; MF: protein kinase activator activity, R-SMAD binding | -0.809 | |
| IRF2BP2 | 359948 | interferon regulatory factor 2 binding protein 2 | BP: transcription, DNA-templated, regulation of transcription, DNA-templated; CC: nucleus, cytoplasm; MF: metal ion binding | -0.805 | |
| LOC148430 | 148430 | -0.803 | |||
| LOC641814 | 641814 | -0.779 | |||
| RPS5 | 6193 | ribosomal protein S5 | BP: translation, nuclear-transcribed mRNA catabolic process, nonsense-mediated decay, translational initiation, translational elongation, translational termination, SRP-dependent cotranslational protein targeting to membrane, gene expression, cellular protein metabolic process; CC: small ribosomal subunit, cytosol, focal adhesion, ribonucleoprotein complex, extracellular exosome; MF: RNA binding, structural constituent of ribosome, poly(A) RNA binding | -0.772 | |
| PGRMC1 | 10857 | progesterone receptor membrane component 1 | CC: endoplasmic reticulum membrane, integral component of membrane, nucleolus, extracellular exosome; MF: steroid binding, protein binding, heme binding | -0.771 | |
| ANXA2 | 302 | annexin A2 | BP: negative regulation of catalytic activity; MF: phospholipase inhibitor activity, calcium ion binding, calcium-dependent phospholipid binding, cytoskeletal protein binding | -0.768 | |
| TNFRSF21 | 27242 | tumor necrosis factor receptor superfamily, member 21 | BP: adaptive immune response, apoptotic process, cellular, cellular response to tumor necrosis factor, humoral immune response, myelination, T cell receptor signaling pathway, negative regulation of interleukin-10 secretion, negative regulation of interleukin-13 secretion, negative regulation of interleukin-5 secretion; CC: integral component of plasma membrane, plasma membrane; MF: protein binding | Cytokine-cytokine receptor interaction | -0.767 |
| PALM | 5064 | paralemmin | BP: regulation of cell shape, movement of cell or subcellular component, negative regulation of adenylate cyclase activity, positive regulation of filopodium assembly, cytoskeleton organization, negative regulation of dopamine receptor signaling pathway, protein targeting to plasma membrane; CC: membrane, nucleus, nucleoplasm, plasma membrane, cytoplasmic membrane-bounded vesicle; MF: protein binding, D3 dopamine receptor binding | -0.767 | |
| SLC25A6 | 293 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | BP: apoptotic process, transmembrane transport, protein targeting to mitochondrion, ADP transport, ATP transport, cellular protein metabolic process; CC: nucleus, mitochondrial inner membrane, integral component of membrane; MF: transporter activity, ATP:ADP antiporter activity, protein binding | Calcium signaling pathway, cGMP-PKG signaling pathway | -0.764 |
| LOC652624 | 652624 | -0.760 | |||
| CKLF | 51192 | chemokine-like factor | BP: cell proliferation, neutrophil chemotaxis, secretion by cell, macrophage chemotaxis, lymphocyte chemotaxis; CC: integral component of membrane, extracellular space; MF: chemokine activity | -0.756 | |
| LOC642741 | 642741 | -0.755 | |||
| LOC730187 | 730187 | -0.746 | |||
| SLC9A3R1 | 9368 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 | BP: adenylate cyclase-activating dopamine receptor signaling pathway, negative regulation of cell proliferation, negative regulation of platelet-derived growth factor receptor signaling pathway, negative regulation of phosphatidylinositol 3-kinase signaling, actin cytoskeleton organization, negative regulation of cell migration, negative regulation of sodium:proton antiporter activity, negative regulation of protein kinase B signaling, glutathione transport, cellular protein localization, phospholipase C-activating dopamine receptor signaling pathway, negative regulation of ERK1 and ERK2 cascade, positive regulation of intrinsic apoptotic signaling pathway, negative regulation of phosphatidylinositol 3-kinase signaling; CC: cytoplasm, intracellular membrane-bounded organelle, centrosome; MF: beta-catenin binding, chloride channel regulator activity, phosphatase binding, PDZ domain binding, beta-2 adrenergic receptor binding, dopamine receptor binding, growth factor receptor binding | -0.745 | |
| CES1 | 1066 | carboxylesterase 1 (monocyte/macrophage serine esterase 1), transcript variant 5 | BP: metabolic process, epithelial cell differentiation; CC: endoplasmic reticulum lumen; MF: methylumbelliferyl-acetate deacetylase activity, carboxylic ester hydrolase activity | Metabolic pathways | -0.745 |
| LOC646785 | 646785 | -0.720 | |||
| TSPAN4 | 7106 | tetraspanin 4 | BP: protein complex assembly; CC: plasma membrane, focal adhesion; MF: antigen binding, integrin binding | -0.718 | |
| SSR2 | 6746 | signal sequence receptor, beta (translocon-associated protein beta) | BP: chromatin remodeling, regulation of transcription from RNA polymerase II promoter, translation, SRP-dependent cotranslational protein targeting to membrane, gene expression, cellular protein metabolic process; CC: endoplasmic reticulum, integral component of membrane, phagocytic vesicle, SWI/SNF complex, RSC complex, cytoplasm; MF: DNA binding, zinc ion binding | Protein processing in endoplasmic reticulum | -0.715 |
| RPL12 | 6136 | ribosomal protein L12 | BP: nuclear-transcribed mRNA catabolic process, nonsense-mediated decay, translation, translational initiation, translational elongation, translational termination, SRP-dependent cotranslational protein targeting to membrane, gene expression, cellular protein metabolic process; CC: cytosol, focal adhesion, cytosolic large ribosomal subunit, extracellular exosome; MF: structural constituent of ribosome, protein binding, poly(A) RNA binding | Ribosome | -0.709 |
| MS4A7 | 58475 | membrane-spanning 4-domains, subfamily A, member 7 | CC: integral component of membrane | -0.709 | |
| TSPO | 706 | translocator protein | BP: positive regulation of apoptotic process, positive regulation of necrotic cell death, negative regulation of nitric oxide biosynthetic process, positive regulation of reactive oxygen species metabolic process, regulation of oxidative phosphorylation, positive regulation of mitochondrial depolarization, negative regulation of tumor necrosis factor production, steroid biosynthetic process, chloride transport, positive regulation of calcium ion transport; CC: mitochondrial envelope, mitochondrial outer membrane, integral component of membrane | Neuroactive ligand-receptor interaction | -0.708 |
| LOC731096 | 731096 | -0.705 | |||
| LOC387930 | 387930 | -0.704 | |||
| PPDPF | 79144 | pancreatic progenitor cell differentiation and proliferation factor | BP: multicellular organismal development, cell differentiation | -0.700 | |
| LOC441013 | 441013 | -0.699 | |||
| LOC647436 | 647436 | -0.699 | |||
| ATF5 | 22809 | activating transcription factor 5 | BP: regulation of transcription from RNA polymerase II promoter, transcription from RNA polymerase II promoter, negative regulation of nucleic acid-templated transcription, negative regulation of apoptotic process; CC: nucleoplasm, cytoplasm, transcription factor complex; MF: sequence-specific DNA binding transcription factor activity, RNA polymerase II transcription regulatory region sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription, transcription corepressor activity, protein binding, heat shock protein binding | -0.694 | |
| APBB1IP | 54518 | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | BP: signal transduction, positive regulation of cell adhesion; CC: cytoplasm, cytoskeleton, plasma membrane, cell junction | Rap1 signaling pathway, Platelet activation | -0.691 |
| LMCD1 | 29995 | LIM and cysteine-rich domains 1 | BP: positive regulation of calcineurin-NFAT signaling cascade, negative regulation of nucleic acid-templated transcription, regulation of cardiac muscle hypertrophy, activation of mitophagy in response to mitochondrial depolarization, negative regulation of transcription from RNA polymerase II promoter, transcription, DNA-templated; CC: nucleus, extracellular space, extracellular matrix; MF: transcription corepressor activity, zinc ion binding | -0.683 | |
| LOC648294 | 648294 | -0.680 | |||
| AVPI1 | 60370 | arginine vasopressin-induced 1 | BP: activation of MAPK activity, cell cycle; MF: protein binding | -0.669 | |
| LOC729679 | 729679 | -0.665 | |||
| LOC645387 | 645387 | -0.665 | |||
| ENSA | 2029 | endosulfine alpha | BP: G2/M transition of mitotic cell cycle, mitotic cell cycle, transport, mitotic nuclear division, regulation of protein phosphatase type 2A activity, negative regulation of catalytic activity, cell division; CC: nucleoplasm, cytoplasm; MF: receptor binding, ion channel inhibitor activity, protein phosphatase type 2A regulator activity, phosphatase inhibitor activity, potassium channel inhibitor activity, protein phosphatase 2A binding | -0.662 | |
| LOC728128 | 728128 | -0.652 | |||
| BTG3 | 10950 | BTG family, member 3 | BP: negative regulation of cell proliferation, negative regulation of mitotic cell cycle; CC: cytoplasm; MF: protein binding | RNA degradation | -0.650 |
| UNG | 7374 | uracil-DNA glycosylase | BP: negative regulation of apoptotic process, DNA repair, base-excision repair, depyrimidination; CC: mitochondrion, nucleus, nucleoplasm; MF: uracil DNA N-glycosylase activity, protein binding | -0.647 | |
| CNN2 | 1265 | calponin 2 | BP: actomyosin structure organization, positive regulation of gene expression, negative regulation of cell migration, regulation of cell proliferation, cytoskeleton organization, cellular response to mechanical stimulus, regulation of actin filament-based process; CC: cytoskeleton, stress fiber, cell-cell junction, focal adhesion, extracellular exosome; MF: actin binding, calmodulin binding | -0.641 | |
| GJC2 | 57165 | gap junction protein, gamma 2 | BP: cell death, cell-cell signaling, response to toxic substance, transmembrane transport; CC: connexon complex, integral component of membrane; MF: gap junction channel activity | -0.639 | |
| PGLS | 25796 | 6-phosphogluconolactonase | BP: carbohydrate metabolic process, pentose-phosphate shunt; CC: cytosol, extracellular vesicular exosome; MF: 6-phosphogluconolactonase activity, monosaccharide binding | Carbon metabolism, Pentose phosphate pathway, Metabolic pathways | -0.639 |
| AGTR1 | 185 | angiotensin II receptor, type 1 | BP: angiotensin-activated signaling pathway, Rho protein signal transduction, positive regulation of cholesterol esterification, regulation of vasoconstriction, calcium-mediated signaling, positive regulation of cellular protein metabolic process, positive regulation of phospholipase A2 activity, positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway, phospholipase C-activating angiotensin-activated signaling pathway, regulation of cell growth; CC: plasma membrane, integral component of membrane; MF: angiotensin type II receptor activity, angiotensin type I receptor activity, bradykinin receptor binding | Vascular smooth muscle contraction, Renin-angiotensin system, Calcium signaling pathway, cGMP-PKG signaling pathway, Neuroactive ligand-receptor interaction, Adrenergic signaling in cardiomyocytes | -0.636 |
| LOC729798 | 729798 | -0.635 | |||
| CAMK1 | 8536 | calcium/calmodulin-dependent protein kinase I | BP: cell cycle, positive regulation of muscle cell differentiation, positive regulation of protein export from nucleus, protein phosphorylation, regulation of protein binding, regulation of protein localization, signal transduction, nucleocytoplasmic transport; CC: cytoplasm, nucleus; MF: ATP binding, protein serine/threonine kinase activity, calmodulin binding, calmodulin-dependent protein kinase activity, protein binding | -0.627 | |
| APRT | 353 | adenine phosphoribosyltransferase | BP: purine-containing compound salvage, purine ribonucleoside salvage, nucleoside metabolic process; CC: cytoplasm; MF: adenine phosphoribosyltransferase activity, hypoxanthine phosphoribosyltransferase activity, AMP binding, transferase activity, transferring glycosyl groups | Purine metabolism, Metabolic pathways | -0.625 |
| SFRS7 | 6432 | splicing factor, arginine/serine-rich 7 | BP: mRNA processing, mRNA export from nucleus, RNA splicing, mRNA splicing, via spliceosome, transcription from RNA polymerase II promoter, termination of RNA polymerase II transcription, negative regulation of mRNA splicing, via spliceosome, gene expression, mRNA 3'-end processing; CC: nucleoplasm, cytoplasm, extracellular exosome; MF: nucleic acid binding, zinc ion binding, nucleotide binding, poly(A) RNA binding, protein binding | -0.624 | |
| LOC653232 | 653232 | -0.618 | |||
| TUBB | 203068 | tubulin, beta | BP: metabolic process, spindle assembly, protein polymerization; CC: nuclear envelope lumen, cytoplasmic ribonucleoprotein granule, extracellular exosome; MF: GTP binding, structural constituent of cytoskeleton, GTPase activity, ubiquitin protein ligase binding | Phagosome, Gap junction | -0.617 |
| ERGIC3 | 51614 | ERGIC and golgi 3 | BP: vesicle-mediated transport; CC: endoplasmic reticulum membrane, Golgi apparatus, endoplasmic reticulum-Golgi intermediate compartment membrane | -0.617 | |
| LOC644315 | 644315 | -0.608 | |||
| RPL26 | 6154 | ribosomal protein L26 | BP: translation, nuclear-transcribed mRNA catabolic process, nonsense-mediated decay, rRNA processing, translational initiation, translational elongation, translational termination, SRP-dependent cotranslational protein targeting to membrane, gene expression, cellular protein metabolic process; CC: large ribosomal subunit, cytosol, extracellular exosome; MF: structural constituent of ribosome, RNA binding, poly(A) RNA binding | Ribosome | -0.604 |
| DHRS3 | 9249 | dehydrogenase/reductase (SDR family) member 3 | BP: oxidation-reduction process, cardiac septum morphogenesis, retinoid metabolic process; CC: endoplasmic reticulum membrane; MF: oxidoreductase activity | Metabolic pathways | -0.603 |
| SEC11C | 90701 | SEC11 homolog C | BP: signal peptide processing, proteolysis, translation, SRP-dependent cotranslational protein targeting to membrane, gene expression, cellular protein metabolic process; CC: integral component of membrane, endoplasmic reticulum membrane; MF: serine-type peptidase activity, hydrolase activity | Protein export | -0.600 |
| LOC643358 | 643358 | -0.598 | |||
| TIGA1 | 114915 | TIGA1 | -0.598 | ||
| LOC285900 | 285900 | -0.596 | |||
| EIF4A1 | 1973 | eukaryotic translation initiation factor 4A, isoform 1 | BP: metabolic process; CC: extracellular vesicular exosome, nucleus, cytoplasm; MF: nucleic acid binding, helicase activity, ATP-dependent helicase activity, ATP binding, double-stranded RNA binding, translation initiation factor activity, poly(A) RNA binding | RNA transport | -0.591 |
| LOC388720 | 388720 | -0.587 | |||
| ADH1C | 126 | alcohol dehydrogenase 1C (class I), gamma polypeptide | BP: oxidation-reduction process, small molecule metabolic process; CC: cytosol; MF: alcohol dehydrogenase (NAD) activity, oxidoreductase activity, zinc ion binding | Metabolic pathways, Glycolysis / Gluconeogenesis, Fatty acid degradation, Tyrosine metabolism, Drug metabolism, Retinol metabolism | -0.587 |
| SEC61B | 10952 | Sec61 beta subunit | BP: intracellular protein transport, protein import into nucleus, translocation, antigen processing and presentation of peptide antigen via MHC class I, translation, SRP-dependent cotranslational protein targeting to membrane, gene expression, ER-associated ubiquitin-dependent protein catabolic process, endoplasmic reticulum unfolded protein response, retrograde protein transport, ER to cytosol, IRE1-mediated unfolded protein response, cellular protein metabolic process; CC: Sec61 translocon complex, endoplasmic reticulum, cytosol, endoplasmic reticulum Sec complex; MF: protein binding, poly(A) RNA binding, epidermal growth factor binding | Protein processing in endoplasmic reticulum, Protein export, Phagosome | -0.583 |
BP: biological process; CC: cell component; MF: molecular function.
Fig 1. Unsupervised hierarchical clustering of RNA microarray expression values.
A total of 112 genes were identified to be differentially up-regulated and 132 genes were identified to be differentially down-regulated in the left atria between mitral regurgitation (MR) patients (n = 7) and normal subjects (NC) (n = 3) by using genefilter R package with the P value < 0.01 (t-test) and a fold-change cut-offs of > 1.5. Bar color indicates mRNA expression level. Red indicates up-regulation; black, no change; green, down-regulation.
To elucidate the molecular mechanisms of MR on left atrial gene expression, we used Ingenuity Pathway Analysis to search for enrichment in predicted function. A network with highest score (P-score = 50, i.e. P value < 10−50) was generated from 244 differentially expressed genes using Ingenuity Pathway Analysis Global Molecular Network algorithm as depicted in Fig 2, and 26 focused genes were identified to be involved in the network, including PLCE1, PPP3R1, PPP3CB, MEF2C and etc. Top involved canonical pathways in this network included role of nuclear factor of activated T cells (NFAT) in cardiac hypertrophy, cardiac hypertrophy signaling, and calcium signaling (Table 5). Top diseases and functions in this network included cardiovascular system development and function, organ morphology, and organismal development (Table 5). These results demonstrated that the network was significantly associated with cardiac related pathways and functions, such as role of NFAT in cardiac hypertrophy and cardiovascular system development and function.
Fig 2. The network with highest score (P-score = 50, i.e. P-value < 10−50) was derived from 244 differentially expressed genes using Ingenuity Pathway Analysis Global Molecular Network algorithm.
The edge in this network represents a relationship between two genes based on Ingenuity Pathways Knowledge Base. The genes with violet border color represent its functions related to cardiovascular system development, such as MEF2C, PLCE1, PPP3CB, and PPP3R1.
Table 5. Top Involved Canonical Pathways and Top Diseases and Functions in the Network Derived from 244 Differentially Expressed Genes between Mitral Regurgitation Patients and Normal Subjects Using Ingenuity Pathway Analysis Global Molecular Network Algorithm.
| Top Canonical Pathways and Diseases and Functions [Genes] | P Value |
|---|---|
| Role of NFAT in cardiac hypertrophy [PPP3R1, PPP3CB, Calcineurin protein(s), PLCE1, Hdac, ERK1/2, Akt, Pp2b, CAMK1, MEF2C] | 4.03E-06 |
| Cardiac hypertrophy signaling [PPP3R1, PPP3CB, Calcineurin protein(s), PLCE1, ERK1/2, Akt, Pp2b, MEF2C] | 2.33E-04 |
| Calcium signaling [PPP3R1, PPP3CB, Calcineurin protein(s), Hdac, ERK1/2, Pp2b, CAMK1, MEF2C] | 8.64E-05 |
| Cardiovascular system development and function [PPP3R1, PPP3CB, Calcineurin protein(s), PLCE1, Hdac, DOCK1, DHRS3, PICALM, ERK1/2, Akt, RGS5, MEF2C] | 3.1E-6–8.08E-2 |
| Organ morphology [PPP3R1, PPP3CB, Calcineurin protein(s), PLCE1, Hdac, DOCK1, DHRS3, PICALM, ERK1/2, Akt, INPP5E, MEF2C] | 3.1E-6–4.67E-2 |
| Organismal development [PPP3R1, PPP3CB, Calcineurin protein(s), PLCE1, Hdac, DOCK1, DHRS3, PICALM, INPP5E, SLC9A3R1, MEF2C] | 3.1E-6–9.22E-2 |
NFAT = nuclear factor of activated T cells.
Furthermore, we applied the activation z-score analysis method, which was proposed by Andreas Krämer et al [5] in 2014, to measure activation states (increased or decreased) of the pathways affected by differentially expressed genes. We take a statistical approach by defining a quantity (z‐score) that determines whether a biological function has significantly more “increased” predictions than “decreased” predictions (z>0) or vice versa (z<0). In practice, z‐scores greater than 2 or smaller than ‐2 can be considered significant. Only “Role of NFAT in cardiac hypertrophy” pathway (Fig 3) had a z-score of 1.34 and P < 0.02. Thus, according to log2 fold-change values and the predictive activities of the differentially expressed genes significantly involved in NFAT pathway (Table 6), we derived an activation z-score equal to 1.34 which suggests that these differentially expressed genes moderately activate the Role of NFAT in cardiac hypertrophy pathway (Fig 3). The detailed information of differentially expressed genes involved in NFAT pathway was shown in Table 6, implicating that the expression patterns of PPP3CB (Calcineurin A beta), PPP3R1 (Calcineurin B), PLCE1, MEF2C, and CAMK1 played a role in the activation of hypertrophy of atrial myocytes in MR patients compared to normal subjects. The results of the real-time quantitative RT-PCR of these 5 genes were consistent with the RNA microarray data (Table 7).
Fig 3. The “Role of NFAT in cardiac hypertrophy” pathway.
Table 6. Log2 Fold Change Values and Predictive Activity of the Differentially Expressed Genes Significantly Involved in Role of NFAT in Cardiac Hypertrophy Pathway.
| Symbol | Entrez Gene Name | Log2FC value | Predictive Activity to Pathway (IPA Knowledge Base) |
|---|---|---|---|
| CAMK1 | calcium/calmodulin dependent protein kinase I | -0.627 | Inhibition |
| MEF2C | myocyte enhancer factor 2C | 0.767 | Activation |
| PLCE1 | phospholipase C epsilon 1 | 1.333 | Activation |
| PPP3CB | protein phosphatase 3 catalytic subunit beta | 0.581 | Activation |
| PPP3R1 | protein phosphatase 3 regulatory subunit B, alpha | -1.267 | Activation |
NFAT = nuclear factor of activated T cells; IPA = Ingenuity Pathway Analysis.
Table 7. Analysis of mRNA Levels via Quantitative RT-PCR and RNA Microarray.
| Gene Name | MR | NC | P Value |
|---|---|---|---|
| PPP3CB (P) | 14.45±0.35 | 16.02±0.19 | 0.007 |
| PPP3CB (M) | 1564.31±86.50 | 1039.52±45.45 | 0.017 |
| PLCE1 (P) | 15.68±0.42 | 17.16±0.42 | 0.079 |
| PLCE1 (M) | 971.23±49.64 | 383.49±19.73 | 0.017 |
| CAMK1 (P) | 18.57±0.24 | 17.43±0.43 | 0.043 |
| CAMK1 (M) | 166.52±6.75 | 256.12±7.70 | 0.017 |
| PPP3R1 (P) | 14.69±0.42 | 13.06±0.27 | 0.017 |
| PPP3R1 (M) | 253.76±29.68 | 595.10±97.05 | 0.017 |
| MEF2C (P) | 15.67±0.56 | 17.82±0.36 | 0.021 |
| MEF2C (M) | 638.07±63.74 | 366.23±29.81 | 0.017 |
Data are presented as mean ± SEM.
(P) = quantitative RT-PCR values (presented in △Cq units). MR (n = 14); NC (n = 6).
(M) = RNA microarray values (presented in normalized fluorescent intensity units). MR (n = 7); NC (n = 3).
Additionally, Significant Analysis of Microarrays method [7] was also applied for significant gene analysis. One thousand and eighty-three significant genes were identified with false discovery rate of 0.05, and four out of five focused genes (PLCE1, CAMK1, PPP3R1, and PPP3CB) were also identified using Significant Analysis of Microarrays method. The canonical pathway of Role of NFAT in Cardiac Hypertrophy was still significantly identified using Ingenuity Pathway Analysis with P-value of 0.039 and z-score of 0.83. Therefore, we focused on deciphering and discussing their regulatory roles in cardiac hypertrophy in the following sections and 5 focused NFAT associated genes (PLCE1, PPP3R1, PPP3CB, CAMK1, MEF2C) were studies for experimental validation.
Hypertrophy of Atrial Myocytes in MR Patients Compared to Normal Subjects and Patients with Aortic Valve Disease
The average cell surface area of myocytes in the left atrial tissue of the MR patients (n = 14) significantly exceeded the average cell surface area of myocytes in the left atrial tissue of the patients with aortic valve disease (n = 5) (1145.3±97.1 vs. 637.7±95.7 μm2, P = 0.012) and normal control subjects (n = 3; 20-year-old Asian male and 48-year-old Asian male, purchased from Abcam, Cambridge, UK and 24-year-old Asian male, purchased from BioChain, Newark, CA, USA) (1145.3±97.1 vs. 491.7±60.7 μm2, P = 0.008) (Fig 4). The average nuclear size of myocytes in the left atrial tissues of the MR patients significantly exceeded the average nuclear size of myocytes in the left atrial tissue of the patients with aortic valve disease (198.8±12.0 vs. 135.7±19.8μm2, P = 0.026) and normal control subjects (198.8±12.0 vs. 129.9±15.1 μm2, P = 0.023) (Fig 4). However, the average cell surface area and nucleus size of myocytes in the left atrial tissue did not significantly differ between patients with aortic valve disease and normal subjects (P = 0.456 and P = 0.881, respectively).
Fig 4.
Histochemical study of hematoxylin, eosin (400 X) stained left atrial tissue sections of (A) mitral regurgitation (MR) patients (n = 14), (B) patients with aortic valve disease (AVD) (n = 5), and (C) normal control (NC) subjects (n = 3). The average cell surface area (D) and average nucleus size (E) per myocyte in the left atrial tissues of MR patients, patients with AVD, and normal control. *P < 0.05. Bar = 50 μm.
α-Sarcomeric Actin Expression in the Left Atria between MR Patients and Normal Subjects
Four normal adult left atrial tissue samples (66-year-old Caucasian female, 49-year-old Africa American male, 62-year-old Asian female and 78-year-old Caucasian female,) were purchased from BioChain, Newark, CA, USA, and these 4 normal atrial tissues were used as the normal controls for protein analysis.
The expression of α-sarcomeric actin protein (normalized against GAPDH) in the left atrial free wall was significantly up-regulated in the MR patients (n = 10) compared to normal subjects (n = 4) (1.30± 0.07 vs. 0.67± 0.13, P = 0.007).
Comparison of the Gene Expression in the “Role of NFAT in Cardiac Hypertrophy” Pathway in the Left Atrium among MR Patients, Patients with Aortic Valve disease and Normal Subjects
The expressions of mRNAs of PPP3CB (MR, n = 13, normal subjects, n = 6; 14.45±0.35 vs. 16.02±0.19, P = 0.007) and MEF2C (MR, n = 14, normal subjects, n = 5; 15.67±0.56 vs. 17.82±0.36, P = 0.021) in the left atrial free wall were significantly up-regulated in the MR patients compared to normal subjects (Fig 5). However, the expressions of mRNAs of CAMK1 (MR, n = 14, normal subjects, n = 6; 18.57±0.24 vs. 17.43±0.43, P = 0.043) and PPP3R1 (MR, n = 14, normal subjects, n = 6; 14.69±0.42 vs. 13.06±0.27, P = 0.017) in the left atrial free wall were significantly down-regulated in the MR patients compared to normal subjects.
Fig 5.
Quantitative determination of mRNAs of (A) protein phosphatase 3, catalytic subunit, beta isozyme (PPP3CB), (B) phospholipase C, epsilon 1 (PLCE1), (C) calcium/calmodulin-dependent protein kinase I (CAMK1), (D) protein phosphatase 3, regulatory subunit B, alpha (PPP3R1), and (E) myocyte enhancer factor 2 (MEF2C) by real-time RT-PCR in the left atrial tissues of mitral regurgitation (MR) patients, patients with aortic valve disease (AVD), and normal control (NC) subjects. *P < 0.05.
The expression of mRNAs of PPP3CB (MR, n = 13, aortic valve disease, n = 6; 14.45±0.35 vs. 17.13±0.49, P = 0.001), MEF2C (MR, n = 14, aortic valve disease, n = 7; 15.67±0.56 vs. 17.37±0.34, P = 0.037) and PLCE1 (MR, n = 13, aortic valve disease, n = 7; 15.68±0.42 vs. 17.10±0.34, P = 0.043) in the left atrial free wall was significantly up-regulated in the MR patients compared to patients with aortic valve disease (Fig 5). However, the expression of mRNAs of CAMK1 (MR, n = 14, aortic valve disease, n = 7; 18.57±0.24 vs. 18.89±0.23, P = 0.502) and PPP3R1 (MR, n = 14, aortic valve disease, n = 7; 14.69±0.42 vs. 13.95±0.21, P = 0.502) in the left atrial free wall did not significantly differ between MR patients and patients with aortic valve disease. These findings implicated that PPP3CB, MEF2C and PLCE1 were associated with the hypertrophy of atrial myocytes in MR patients compared to patients with aortic valve disease.
The expressions of mRNAs of CAMK1 (aortic valve disease, n = 7, normal subjects, n = 6; 18.89±0.23 vs. 17.43±0.43, P = 0.032) and PPP3R1 (aortic valve disease, n = 7, normal subjects, n = 6; 13.95±0.21 vs. 13.06±0.27, P = 0.032) in the left atrial free wall were significantly down-regulated in patients with aortic valve disease compared to normal subjects (Fig 5). However, there was no significant difference in the expressions of mRNAs of PPP3CB (aortic valve disease, n = 6, normal subjects, n = 6; 17.13±0.49 vs. 16.02±0.19, P = 0.150), PLCE1 (aortic valve disease, n = 7, normal subjects, n = 6; 17.10±0.34 vs. 17.16±0.42, P = 0.886) and MEF2C (aortic valve disease, n = 7, normal subjects, n = 5; 17.37±0.34 vs. 17.82±0.36, P = 0.685) in the left atrial free wall between patients with aortic valve disease and normal subjects.
Comparison of the Expressions of miR-1, miR-133 and miR-208 in the Left Atrium between MR Patients and Normal Subjects
The expression of miR-1 in the left atrial tissue was significantly down-regulated in the MR patients (n = 5) compared to normal controls (n = 5) (-2.794±0.561 vs. -5.252±0.807, P = 0.047). The expression of miR-133 in the left atrial tissue was significantly down-regulated in the MR patients (n = 5) compared to normal controls (n = 5) (-0.065±0.334 vs. -2.083±0.691, P = 0.028). The expression of miR-208 in the left atrial tissue was significantly up-regulated in the MR patients (n = 5) compared to normal controls (n = 5) (-0.995±0.415 vs. 1.460±0.918, P = 0.028).
Hypertrophy of HL-1 Atrial Myocytes by Mechanical Stretching
The causal relationship between atrial hypertrophy and atrial dilatation due to volume overload of MR was partly mimicked by mechanical stretching of HL-1 atrial myocytes. The average cell surface area of stretched HL-1 atrial myocytes (experiment number = 6) significantly exceeded that of non-stretched control group (experiment number = 6) (1203.6±89.0 vs. 819.4±43.1 μm2, P = 0.016). The average nucleus size of stretched HL-1 atrial myocytes significantly exceeded that of non-stretched control group (190.2±18.9 vs. 108.3±8.6 μm2, P = 0.009) (Fig 6).
Fig 6. Immunofluorescence study of average cell surface area and average nucleus size of HL-1 atrial myocytes between the stretched group and non-stretched control group.
Myocyte identification was performed with Phalloidin F-actin (green color). Nucleus identification was performed with Hoechst 33258 (blue color). *P < 0.05. Bar = 80 μm.
bgThe gene expressions of CAMK1 (4.22±0.07 vs. 4.01±0.06, P = 0.037) and PPP3R1 (3.89±0.08 vs. 3.09±0.06, P = 0.004) (normalized against GAPDH) in the HL-1 atrial myocytes were significantly down-regulated in the stretched HL-1 atrial myocytes compared to the non-stretched control group.
Discussion
This study identifies and reports the alteration of the RNA expression pattern, molecular mechanisms and biological processes involving in atrial myocyte hypertrophy between the left atrial myocardium of MR patients and normal subjects using high-density oligonucleotide microarrays and enrichment analysis. A total of 112 genes were identified to be differentially up-regulated and 132 genes were identified to differentially down-regulated in the left atria between MR patients and normal subjects. Notably, the expression patterns of PPP3CB, PPP3R1, PLCE1, MEF2C and CAMK1 in the “NFAT in cardiac hypertrophy” pathway played a significant role in the activation of hypertrophy of atrial myocytes in MR patients compared to normal subjects.
Activation of “Role of NFAT in Cardiac Hypertrophy” Pathway
Calcineurin/NFAT coupling has been reported to participate in pathological, but not physiological, cardiac hypertrophy [8]. Cardiac hypertrophy is a compensatory response to pathological states and hemodynamic overload. However, pathological hypertrophy leads to atrial myocardial disarrangement and consequently, atrial enlargement that is correlated with poor prognosis in MR patients [2].
The initial phase in the development of myocardial hypertrophy involves factors, such as endothelin-1, angiotensin-II, and adrenergic agonists at the cell membrane, binding to the G-protein coupled receptors (Fig 3), and several interdependent signaling cascades that include G-proteins, GTPases such as Ras, RhoA and Rac, and kinases such as ERK/MAPK and PKC [9]. Prior study showed enhanced expression of Rho-associated kinase in the left atrial myocytes of MR patients [10]. In all of the hypertrophic pathways, NFAT plays a critical role in the development of cardiac hypertrophy. Several studies have shown the importance of Ca2+ sensitive signaling molecules, including calcineurins (i.e. PPP3CB, PPP3R1) and CAMK, a calcium/calmodulin-dependent protein kinase, in hypertrophic pathways [11]. Activation of protein kinase C leads to increased Ca2+ levels that activate calcineurins. Calcineurin activation leads to the dephosphorylation of NFATc4, allowing its nuclear localization where it cooperates with other transcription factors to participate in the cardiac hypertrophy. In this study, the expression of PPP3CB, the catalytic subunit, was significantly up-regulated in the MR patients compared to normal subjects. However, the expression of PPP3R1, the regulatory subunit B, was significantly down-regulated in the MR patients compared to normal subjects. Overexpression of PPP3R1, also known as modulatory calcineurin-interacting protein-1, has been reported to attenuate left ventricular hypertrophy after myocardial infarction [12]. As shown in Table 6, the predictive activities of the differential trend between PPP3CB and PPP3R1 to the Role of NFAT in cardiac hypertrophy pathway implicated activation of this pathway. The CAMK1 was also found to be significantly down-regulated in MR patients compared to normal subjects in this study. However, the molecular function of CAMK1 in human heart is rarely reported. Further studies are warranted to investigate the role of CAMK1 in the development of atrial hypertrophy in patients with MR.
MEF2 (myocyte enhancer factor 2), especially MEF2C, is an important transcription factor regulating the cardiac gene program during myocardial cell hypertrophy [13]. The activation of MEF2 by CAMK is mediated mainly through the phosphorylation of transcriptional repressors, the class II histone deacetylases, resulting in disrupting the association between MEF2 and histone deacetylases in the nucleus and transcriptional activation of cardiac hypertrophy [14,15]. In this study, MEF2C was found to be significantly up-regulated in MR patients compared to normal subjects.
PLCE (phospholipase C epsilon), an effector of Ras, has been shown to involve in stress-induced hypertrophy and PLCE, scaffolded to muscle-specific A kinase-anchoring protein in cardiac myocytes, responds to hypertrophic stimuli to generate diacylglycerol from phosphatidylinositol 4-phosphate in the Golgi apparatus, in close proximity to the nuclear envelope, to regulate activation of nuclear protein kinase D and hypertrophic signaling pathways [16]. In this study, PLCE1 was found to be up-regulated in MR patients compared to patients with aortic valve disease and normal subjects.
Hypertrophy-Related MicroRNAs
The expressions of antihypertrophic miRs, miR-1 and miR-133, in the left atrial tissue were significantly down-regulated in the MR patients compared to normal controls, while the expression of agonist of the hypertrophic response, miR-208, in the left atrial tissue were significantly up-regulated in the MR patients compared to normal controls [17].
Hypertrophy-Related Decorin and Calponin
Decorin has been reported to promote myoblast proliferation mediated by an endoplasmic reticulum stress-related pathway [18]. Decorin was identified to be differentially down-regulated in MR patients compared to normal subjects in this study. Therefore, decorin might not be involved in the hypertrophy of atrial myocytes in MR patients, which was mainly related to the Role of NFAT in cardiac hypertrophy pathway.
Calponin has been reported to be involved in the hypertrophy of smooth muscle cells [19,20]. Calponin 2 was identified to be differentially down-regulated in MR patients compared to normal subjects in this study. Therefore, calponin 2 might not be involved in the hypertrophy of atrial myocytes in MR patients.
Pathological Hypertrophy of Atrial Myocytes in MR
Atria, like the ventricles, can undergo hypertrophy in response to increased volume and pressure overload. In MR, the volume and pressure in the left atrium are greatly increased. The left atrium of MR patients responds by undergoing chronic dilation, which enables it to accommodate the increased volume without a large increase in pressure because of its increased compliance. However, extreme hypertrophy and dilatation is deleterious because it increases the oxygen demand of the heart and decreases mechanical efficiency. Furthermore, atrial fibrillation, an important risk factor of stroke and systemic embolization, may develop as a consequence of atrial enlargement, and vice versa [21].
Study Limitations
There are several limitations of this study. Firstly, most of the patients with MR received renin-angiotensin system blockers. Therefore, the expression of some genes might have been modified by renin-angiotensin system blockers [22]. However, there was no significant difference in the expressions of PPP3CB (14.66±0.38 vs. 13.28±0.02, P = 0.167), PPP3R1 (14.29±0.41 vs. 16.16±0.91, P = 0.052), PLCE1 (15.95±0.46 vs. 14.22±0.02, P = 0.167), MEF2C (15.99±0.58 vs. 14.46±1.47, P = 0.392) and CAMK1 (18.49±0.29 vs. 18.86±0.38, P = 0.484) between MR patients with renin-angiotensin system blockers (n = 11) vs. MR patients without renin-angiotensin system blockers (n = 3). Secondly, the sample size was relatively small. However, the gene expression pattern by microarray analysis was quite consistent in the same group (Fig 1). Thirdly, the age of the normal subjects (n = 6) was younger than that of MR patients (n = 14) (49±25 vs. 58±9 years, P = 0.620), however the difference did not reach statistical significance. Finally, the microarrays were conducted on frozen, unsorted tissue samples. It is hence impossible to ascertain the sub-tissue or cellular origin of generated data. However, histological analysis did show atrial myocyte hypertrophy of MR patients compared to patients with aortic valve disease and normal subjects, implicating at least some involvement of cellular origin.
Conclusions
Significant hypertrophy developed in the left atrial myocytes of MR patients compared to normal subjects and patients with aortic valve disease. The differentially expressed genes in the “Role of NFAT in cardiac hypertrophy” pathway may play a critical role in the atrial myocyte hypertrophy of MR patients and these differentially expressed genes may serve as potential targets for human MR to prevent the progression of left atrial enlargement and its related complications, such as atrial fibrillation, and heart failure.
Accession Codes
The data discussed in this manuscript have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE63045 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63045).
Acknowledgments
The authors would like to thank Chang Gung Medical Foundation Kaohsiung Chang Gung Memorial Hospital Tissue Bank Core Lab (CLRPG8B0033 and CLRPG8E0161) for excellent technical support.
Data Availability
1.All relevant data are within the paper and its Supporting Information files 2.Accession Codes: The data discussed in this manuscript have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE63045 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63045).
Funding Statement
This work was supported by grants from the National Science Council, and the Chang Gung Memorial Hospital, Taiwan (grant number NSC 101-2314-B-182A-096, NSC 102-2314-B-182A-107-MY2, CMRPG8A0571). MCC received all the fundings. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Iung B, Gohlke-Bärwolf C, Tornos P, Tribouilloy C, Hall R, Butchart E, et al. Working Group on Valvular Heart Disease. Recommendations on the management of the asymptomatic patient with valvular heart disease. Eur Heart J 2002;23:1253–1266. [DOI] [PubMed] [Google Scholar]
- 2.Otto CM. Timing of surgery in mitral regurgitation. Heart 2003;89:100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corradi D, Callegari S, Maestri R, Ferrara D, Mangieri D, Alinovi R, et al. Differential structural remodeling of the left-atrial posterior wall in patients affected by mitral regurgitation with or without persistent atrial fibrillation: a morphological and molecular study. J Cardiovasc Electrophysiol 2012;23:271–279. 10.1111/j.1540-8167.2011.02187.x [DOI] [PubMed] [Google Scholar]
- 4.Chang JP, Chen MC, Lin WY, Liu WH, Chen CJ, Chen YL, et al. DNA repair in TUNEL-positive atrial cardiomyocytes of mitral and tricuspid valve diseases: potential mechanism for preserving cardiomyocytes. Int J Cardiol 2011;146:44–50. 10.1016/j.ijcard.2009.06.012 [DOI] [PubMed] [Google Scholar]
- 5.Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523–530. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentleman R, Carey V, Huber W and Hahne F. genefilter: genefilter: methods for filtering genes from microarray experiments. R package version 1.46.1.
- 7.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001;98:5116–121. 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkins BJ, Dai YS, Bueno OF, Parsons SA, Xu J, Plank DM, et al. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ Res 2004;94:110–118. 10.1161/01.RES.0000109415.17511.18 [DOI] [PubMed] [Google Scholar]
- 9.Kang M, Chung KY, Walker JW. G-protein coupled receptor signaling in myocardium: not for the faint of heart. Physiology 2007;22:174–184. 10.1152/physiol.00051.2006 [DOI] [PubMed] [Google Scholar]
- 10.Chen HC, Chang JP, Chang TH, Lin YS, Huang YK, Pan KL, et al. Enhanced expression of ROCK in left atrial myocytes of mitral regurgitation: a potential mechanism of myolysis. BMC Cardiovasc Disord 2015;15:33 10.1186/s12872-015-0038-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bueno OF, Wilkins BJ, Tymitz KM, Glascock BJ, Kimball TF, Lorenz JN, et al. Impaired cardiac hypertrophic response in Calcineurin A beta -deficient mice. Proc Natl Acad Sci U S A 2002;99:4586–4591. 10.1073/pnas.072647999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rooij E, Doevendans PA, Crijns HJ, Heeneman S, Lips DJ, van Bilsen M, et al. MCIP1 overexpression suppresses left ventricular remodeling and sustains cardiac function after myocardial infarction. Circ Res 2004;94:e18–26. 10.1161/01.RES.0000118597.54416.00 [DOI] [PubMed] [Google Scholar]
- 13.Kolodziejczyk SM, Wang L, Balazsi K, DeRepentigny Y, Kothary R, Megeney LA. MEF2 is upregulated during cardiac hypertrophy and is required for normal post-natal growth of the myocardium. Curr Biol 1999;9:1203–1206. 10.1016/S0960-9822(00)80027-5 [DOI] [PubMed] [Google Scholar]
- 14.Akazawa H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res 2003;92:1079–1088. 10.1161/01.RES.0000072977.86706.23 [DOI] [PubMed] [Google Scholar]
- 15.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol 2002;22:7302–7312. 10.1128/MCB.22.20.7302-7312.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Malik S, Pang J, Wang H, Park KM, Yule DI, et al. Phospholipase Cε hydrolyzes perinuclear phosphatidylinositol 4-phosphate to regulate cardiac hypertrophy. Cell 2013;153:216–227. 10.1016/j.cell.2013.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Da Costa Martins PA, De Windt LJ. MicroRNAs in control of cardiac hypertrophy. Cardiovasc Res 2012;93:563–572. 10.1093/cvr/cvs013 [DOI] [PubMed] [Google Scholar]
- 18.Sun L, Lu K, Liu H, Wang H, Li X, Yang C, et al. The effects of endoplasmic reticulum stress response on duck decorin stimulate myotube hypertrophy in myoblasts. Mol Cell Biochem 2013;377:151–161. 10.1007/s11010-013-1581-2 [DOI] [PubMed] [Google Scholar]
- 19.di Gioia CR, van de Greef WM, Sperti G, Castoldi G, Todaro N, Ierardi C, et al. Angiotensin II increases calponin expression in cultured rat vascular smooth muscle cells. Biochem Biophys Res Commun 2000;279:965–969. 10.1006/bbrc.2000.4049 [DOI] [PubMed] [Google Scholar]
- 20.McWhinnie R, Pechkovsky DV, Zhou D, Lane D, Halayko AJ, Knight DA, et al. Endothelin-1 induces hypertrophy and inhibits apoptosis in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2007;292:L278–286. 10.1152/ajplung.00111.2006 [DOI] [PubMed] [Google Scholar]
- 21.Sanfilippo AJ, Abascal VM, Sheehan M, Oertel LB, Harrigan P, Hughes RA, et al. Atrial enlargement as a consequence of atrial fibrillation. A prospective echocardiographic study. Circulation 1990;82:792–797. [DOI] [PubMed] [Google Scholar]
- 22.Chen MC, Chang JP, Chang TH, Hsu SD, Huang HD, Ho WC, et al. Unraveling regulatory mechanisms of atrial remodeling of mitral regurgitation pigs by gene expression profiling analysis: role of type I angiotensin II receptor antagonist. Transl Res 2015;165:599–620. 10.1016/j.trsl.2014.11.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
1.All relevant data are within the paper and its Supporting Information files 2.Accession Codes: The data discussed in this manuscript have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE63045 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63045).