Figure 1.
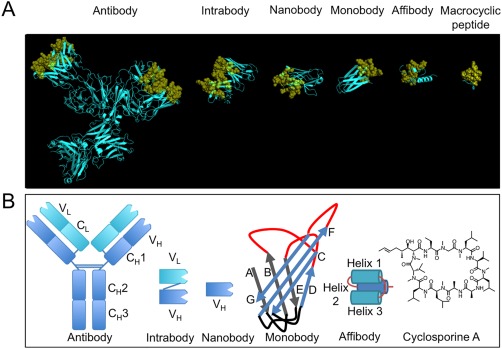
A comparison of ligands with different scaffolds. (A) From left to right, X‐ray crystal structures of: intact murine IgG1 monoclonal antibody for phenobarbital (PDB: 1IGY); single domain intrabody that binds to activated GTP‐bound RAS (PDB: 2UZI); humanized NbBcII10 nanobody that binds to BcII β‐lactamase (PDB: 3EAK); monobody ySMB‐9 that binds to human small ubiquitin‐like modifier 1 (PDB: 3RZW); affibody scaffold Z domain of Staphylococcal protein A (PDB: 1Q2N); macrocyclic peptide aCAP (PDB: 3WMG). Variable binding positions have been coloured yellow and are displayed in cartoon format and semi‐transparent spheres. Structural or non‐varied regions are coloured cyan and are displayed in cartoon format. X‐ray crystal structures were rendered in PyMOL v1.5.0.4. (B) Schematic structures of all ligands discussed, from left to right: monoclonal antibody, intrabody, nanobody, monobody, affibody and a chemical structure of a natural macrocyclic peptide, cyclosporin A
