Figure 1.
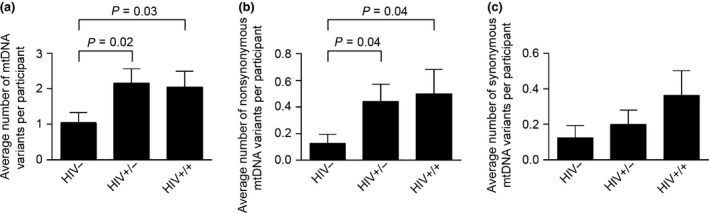
Mitochondrial point mutation frequency among study participants. Mitochondrial DNA (mtDNA) was extracted from peripheral blood mononuclear cells (PBMCs) and amplified with eight pairs of overlapping primers. The PCR products were purified and sequenced with SMRT sequencing technology as described in the ‘Methods’ section. The sequences were demultiplexed and aligned against the revised Cambridge reference sequence for the human mitochondrial genome (NC_012920.1). (a) Average number of mtDNA variants per participant. (b) Average number of nonsynonymous mtDNA variants per participant. (c) Average number of synonymous mtDNA variants per participant. Comparison of synonymous or nonsynonymous changes in the coding region was carried out using the χ2 test. Data are presented as mean ± standard error of the mean. The cut‐off for significance was set at P < 0.05. HIV−, HIV‐uninfected controls; HIV+/−, HIV‐infected treatment‐experienced patients without toxicity; HIV+/+, HIV‐infected treatment‐experienced patients with toxicity.
