Abstract
Jasmonic acid is a plant hormone that can be produced by the fungus Lasiodiplodia theobromae via submerged fermentation. From a biotechnological perspective jasmonic acid is a valuable feedstock as its derivatives serve as important ingredients in different cosmetic products and in the future it may be used for pharmaceutical applications. The objective of this work was to improve the production of jasmonic acid by L. theobromae strain 2334. We observed that jasmonic acid formation is dependent on the culture volume. Moreover, cultures grown in medium containing potassium nitrate as nitrogen source produced higher amounts of jasmonic acid than analogous cultures supplemented with ammonium nitrate. When cultivated under optimal conditions for jasmonic acid production, L. theobromae secreted several secondary metabolites known from plants into the medium. Among those we found 3-oxo-2-(pent-2-enyl)-cyclopentane-1-butanoic acid (OPC-4) and hydroxy-jasmonic acid derivatives, respectively, suggesting that fungal jasmonate metabolism may involve similar reaction steps as that of plants. To characterize fungal growth and jasmonic acid-formation, we established a mathematical model describing both processes. This model may form the basis of industrial upscaling attempts. Importantly, it showed that jasmonic acid-formation is not associated to fungal growth. Therefore, this finding suggests that jasmonic acid, despite its enormous amount being produced upon fungal development, serves merely as secondary metabolite.
Introduction
Due to the limited amount of exhaustable resources and the constantly increasing prices thereof renewable resources are of rising interest [1]. Besides oils and lipids that serve as essential raw material for chemical industry [2], also compounds of the secondary metabolism of different species are becoming an important focus in industrial research. Those compounds are often used as medicinal drugs. In addition, many novel constitutents of cosmetics, pharmaceuticals and nutraceuticals are developed on the physico-chemical basis of the core structures of secondary metabolites [3].
Jasmonates consitute one group of metabolites that is of economical importance. They are α-linolenic acid-derived compounds that exhibit a cyclopentanone ring as structural core-element to which one aliphatic and one carboxylic side chain is attached. The major representatives are jasmonic acid (JA), its methyl ester (MeJA) as well its isoleucine conjugate (JA-Ile). Jasmonates are widely distributed in algae [4], higher plants [5] and microorganisms [6]. Over the past decades a large body of research has been spend on the analysis of JA-function and JA-metabolism in plants and many details are known today. In plants jasmonates play important roles as growth inhibitors, they stimulate plant senescence, and they are also involved in flower development. Furthermore, they function as regulators for plant immunity that induces the expression of defensive genes after pathogen attack or feeding insects [7]. JA biosynthesis is catalyzed in two spatially separated cell compartments–the plastid and the peroxisome. In the former one the biosynthetic pathway is initiated by the peroxidation of α-linolenic acid derived from a plastidial membrane by the action of a 13S-lipoxygenase (13S-LOX). In the following reactions the hydroperoxy fatty acid is converted by allene-oxide synthase (AOS) and allene-oxide cyclase (AOC) to 12-oxo-phytodienoic acid (OPDA). This compound represents the first cyclic intermediate in this pathway. After its transport to the peroxisome, the ring system is reduced and the octanoic side chain is further processed by three rounds of β-oxidation finally yielding JA [8]. Besides this well-studied biosynthesis in plants, neither the function nor the biosynthetic pathways leading to JA in microorganisms are known yet.
MeJA was the first jasmonate that has been identified. It was originally isolated as an odoriferous constituent of the essential oil of Jasminun grandiflorum [9]. This compound is of special interest for perfume and flavor industry as it is an important component for many fragrance mixtures found in cosmetic (i. e. soaps, shampoos) and home care products (i. e. household cleaners) [10]. In the future, MeJA might also become more important for pharmaceutical industry as recent studies suggested that MeJA not only exhibits antidepressant, anti-aggressive activities in mammals but also shows anti-inflammatory effects comparable to that of prostaglandins [11]. Furthermore, growth of some cancerous cells lines of rats and humans can be suppressed by jasmonates in vitro and in vivo [12, 13]. These findings underline the potential use of those compounds as medicinal drugs.
Due to the low concentration necessary for hormonal function in vivo, plants naturally synthesize JA only in very small amounts. This makes the isolation of this compound for industrial purposes difficult and expensive. However, recent studies demonstrated that some fungal species are also capable of producing JAs in amounts exceeding those formed by plants [14–17]. Thus, it can be suggested that exploitation of those systems might be suitable for the industrial production of jasmonates. Different jasmonates have been detected in Lasiodiplodia theobromae (synonym Botryodiplodia theobromae) [18–21], Fusarium oxysporum [22], Aspergillus niger [23] and Gibberella fujikuroi [24]. However, details about biosynthetic routes of fungal JA-biosynthesis are still scarce and it is still unclear whether enzymatic pathways leading to formation of JA are similar to those in plants. Using a reverse genetic approach a specific 13S-LOX has recently been identified in F. oxysporum that might initiate JA-biosynthesis by catalysing the initial oxygenation reaction similar to that known from plants as described above [25]. Potential enzymes acting downstream of 13S-LOX—as for instance AOS or AOC in plants—have not been identified, yet. Hoffmann and co-worker reported, however, on the identification of a novel 9R-dioxygenase-AOS fusion enzyme that oxidizes linoleic acid to its corresponding 9S-hydroperoxy derivative and further isomerizes this intermediate yielding 9S(10)-epoxy-10,12-octadecadienoic acid in F. oxysporum [26]. Interestingly, a similar enzymatic activity has also been found in L. theobromae suggesting related metabolic functions in both fungi [27].
The objective of the present study was on the one hand to optimize the cultivation conditions of L. theobromae in order to increase JA yield and to obtain a kinetic model for fungal growth in respect to JA production. For this purpose, we not only investigated the influence of the different cultivation parameters on JA formation but also quantified the effect on metabolites that might be formed upstream and downstream from JA. On the other hand, a further objective was to identify the main compounds secreted by L. theobromae and thereby to gain more information about fungal secondary metabolism.
Materials and Methods
Microorganism
L. theobromae strain 2334 from the lnstituto Nacional de Investigaciones Fundamentales de la Agricultura Tropical (Cuba) isolated from sunflower, was used. The strain was stored on potato dextrose agar (PDA) slants with glycerol (50%) at 4°C and in mycelial discs on PDA with glycerol (17%) at -80°C.
Culture medium
Culture medium with the following composition was used (in g/L): glucose, 50; KNO3, 8.9; KH2PO4, 2.0; KCl, 0.3; MgSO4∙7H20, 0.6; FeSO4∙7H20, 0.6; ZnSO4∙7H20, 0.03; MnSO4∙7H20, 0.003; CuSO4∙7H20, 0.003; Na2MoO4∙2H20, 0.003; yeast extract, 1.0. Before autoclaving, initial pH was adjusted to 5.5–5.6 with NaOH (0.1 M). The study on the effect of NH4NO3 as nitrogen source was carried out using the same nitrogen concentration (n(N) = 0.088 M) in the culture media as with KNO3 (n(N) = 0.088 M).
Culture techniques
A sample of the stock culture was transferred to PDA plates and incubated for 3 d at 30°C. Five, ten or fifteen disks of mycelium (7 mm diameter) were used for inoculation of 25 mL, 50 mL or 100 mL of culture medium in 100 mL, 250 mL or 500 mL Erlenmeyer flasks, respectively. The medium was supplemented with either NH4NO3 or KNO3 as nitrogen source. Cultures were grown for 12 d in surface cultures at 30°C.
Analytical methods
Biomass concentration was determined by dry weight after broth filtration on cellulose membrane (Rotilabo®-round filters, Type 12A, retention range 16 μm, Carl Roth &Co., Karlsruhe, Germany) followed by drying at 60°C for 24 h. Glucose content was determined by hexokinase method as described [28]. JAs and phytohormone extraction was performed as described previously with some minor modifications [29]. Metabolites were extracted from the culture filtrate at 4°C under constant shaking (200 rpm, 60 min) in the dark using 0.75 mL MeOH and 2.5 mL Methyl-tert-butylether (MTBE). For quantification, deuterated standards were added (50 ng D5-JA, 150 ng D5-OPDA, 50 ng D6-salicylic acid (SA) and 100 ng D5-indole acetic acid (IAA)). Prior to centrifugation 0.6 mL H2O were added to enhance phase separation. The upper phase was collected; the lower phase was re-extracted with 0.7 mL MeOH/H2O (3:2.5, v/v) and 1.3 mL MTBE. The combined phases were evaporated under streaming nitrogen to dryness and re-suspended in a solution of acetonitrile/water/acetic acid (20/80/0.1, v/v/v). The analysis was performed using a 1100 HPLC system (Agilent Technologies, Santa Clara, USA) coupled to a 3200 hybrid triple quadrupole (ABSciex, Framingham, USA) as reported [30]. Mass transitions (in Da) were as follows: 214/62 for D5-JA, 209/59 for JA; 225/59 for 11/12-hydroxy-JA, 237/165 for OPC-4, 296/170 for D5-OPDA, 291/165 for cis-OPDA, 179/135 for D5-IAA, 174/130 for IAA, 160/116 for (indole carboxylic acid) ICA, 137/93 for SA, 141/97 for D6-SA and 207/137 for chorismate.
Structure confirmation of OPC-4 and 11/12-hydroxy-JA was well as structure elucidation of compounds 1–7 were done by high resolution MS/MS fragmentation studies with an ultra-high performance chromatography-electrospray ionization-quadrupole time-of-flight-mass spectrometer (UHPLC-ESI-QTOF-MS). A LC 1290 Infinity (Agilent Technologies, Karlsruhe, Germany) coupled with a 6540 UHD Accurate-Mass Q-TOF LC MS instrument with Jet Stream Technology as ESI source (Agilent Technologies, Karlsruhe, Germany) was used for all analyses. For LC an ACQUITY UPLC HSS T3 column (2.1 x 100 mm, 1.8 μm particle size, Waters Corporation, Milford, USA) was used at a flow rate of 0.5 mL/min at a temperature of 40°C. The solvent system consists of solvent A (water/formic acid (100/0.1, v/v) and solvent B (acetonitrile/formic acid (100/0.1, v/v). The following gradient was applied: 0–3 min from 1% to 20% solvent B, 3–8 min from 20% to 100% solvent B, 8–12 min 100% solvent B. The Q-TOF MS instrument was operated in Extended Dynamic Range and targeted MS/MS mode with a frequency of 2 GHz. Source conditions were: gas temperature: 250°C; drying gas flow: 8 L/min; nebulizer pressure: 35 psi; sheath gas temperature: 300°C; sheath gas flow: 8 L/min; VCap voltage: 3 kV; nozzle voltage: 200 V; fragmentor voltage: 100 V. Samples were ionized in negative and/or positive ESI mode with collision energy 10 eV. Data were acquired by Mass Hunter Workstation Acquisition software B.06 (Agilent Technologies, Karlsruhe, Germany) and analyzed by Mass Hunter Qualitative Analysis B.06 (Agilent Technologies, Karlsruhe, Germany).
Results
Identification of a potential JA precursor and oxidized JA derivatives
Prior to our optimization approach, we were interested in potential metabolites that might be formed up-stream and down-stream of JA in order to gain a deeper understanding of fungal JA-metabolism. Because in plants OPDA and specific 3-oxo-2-(pent-2-enyl)-cyclopentane-1-carboxylic acid (OPC)-derivatives are prominent JA-precursors, we primarily focused on the identification of those compounds. Moreover, we considered that oxidized JA-derivatives may be formed as catabolic JA-products similar to those that have been observed in plants [31–34].
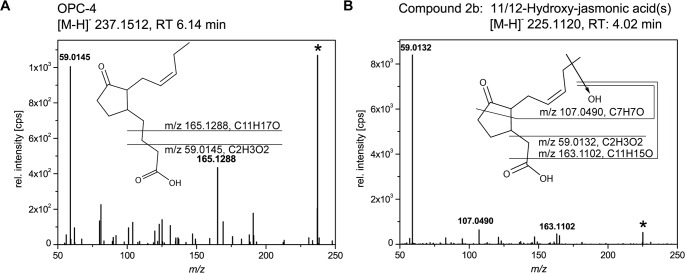
In search for those compounds we analysed the culture filtrate of L. theobromae strain 2334 by using UHPLC-ESI-QTOF-MS. We observed formation of 3-oxo-2-(pent-2-enyl)-cyclopentane-1-butanoic acid (OPC-4) and hydroxy-derivatives of JA (Fig 1), whereas OPDA could only be detected in trace amounts. The chemical structure of OPC-4 was confirmed by retention time and high resolution MS/MS-spectra as the corresponding authentic standard (Fig 1A). The parent ion represents the base peak at m/z 237.1512. Two diagnostic signals were at m/z 59.0145 (carboxy-methyl group) and m/z 165.1288, which corresponds to the sum formula C11H17O- (Fig 1A) [35].
Fig 1. UHPLC-ESI-QTOF-MS analysis of OPC-4 and 11/12-hydroxy-JA formed by L. theobromae strain 2334.
The fungus was cultivated in medium supplemented with KNO3 for 9 days (d). After extraction compounds of the culture filtrate were analyzed via UHPLC-ESI-QTOF-MS. Shown are high resolution MS/MS spectra (negative ionization mode; collision energy 10 eV) of (A) OPC-4 ([M-H]- 237.1512, RT 6.14 min) and (B) 11/12-hydroxy-JA ([M-H]- 225.1120, RT 4.02 min). Proposed fragmentation is shown as inset. Abbreviations: JA, jasmonic acid; OPC-4, 3-oxo-2-(pent-2-enyl)-cyclopentane-1-butanoic acid.
The MS/MS-spectrum of hydroxy-JA (compound 2b) is shown in Fig 1B. This compound exhibited an accurate mass of [M-H]- 225.1120 and eluted at 4.02 min. The respective MS/MS-spectra displayed three fragment ions characteristic for JA-derivatives in the negative ionization mode. The base-peak was found at m/z 59.0132, which corresponds to the m/z of the carboxy-methyl group from the JA-backbone. It has been reported before that formation of this fragment is diagnostic for JA-derivatives [36]. An additional characteristic signal at m/z 163.1102 belongs to the fragment ion of [M-CO2-H2O-H]-, which results from neutral loss of the carboxyl-group as CO2 and the hydroxy-group as water. The signal at m/z 107.0490 arises from a cleavage of the cyclopentanone ring of the JA-backbone and the loss of hydroxy-group in C11 or C12 position yielding a C7H7O—fragment as indicated in Fig 1B. Based on this result, we assigned this metabolite as a hydroxy-JA. In order to evaluate the position of the hydroxy-group we used authentic 11-hydroxy- and 12-hydroxy-JA as standard. Both compounds eluted at the same retention time as compound 2b and gave the identical MS/MS-spectra suggesting that L. theobromae might either form 11-hydroxy, 12-hydroxy- or even both hydroxylated JA isomers. 11- and 12-hydroxy-JAs were described by Miersch and co-workers [19]. For this reason, we termed this peak 11/12-hydroxy-JA.
Influence of cultivation conditions on fungal growth, production of JA, OPC-4, and 11/12-hydroxy-JA
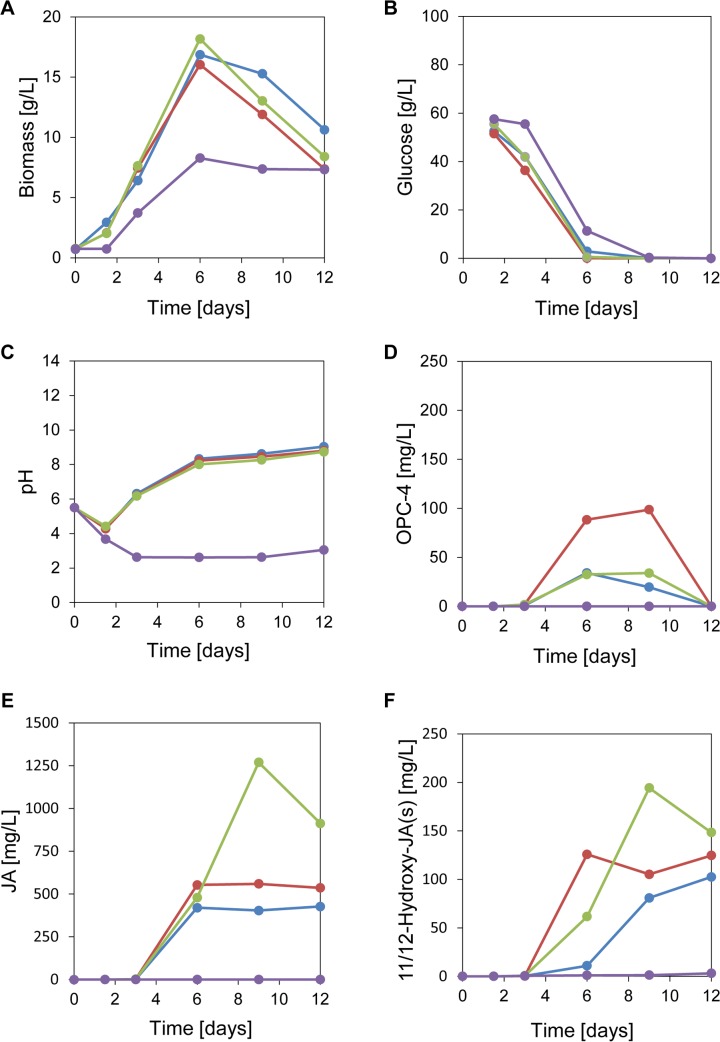
Under static culture conditions L. theobromae forms a mat on the surface of the culture medium [14]. Therefore, we started our optimization approach by investigating the influence of the culture volume and the associated surface area on biomass-production, glucose consumption, pH-changes and JA-metabolism. For this purpose, we cultivated L. theobromae strain 2334 in shake flasks of different sizes (100 mL, 250 mL and 500 mL) using culture medium supplemented with KNO3 as nitrogen source and monitored the different parameters mentioned above. In order to evaluate the influence of the nitrogen source on fungal growth and JA-metabolism, we performed additional experiments using culture medium that was supplemented with NH4NO3 instead of KNO3. The results are presented in Fig 2.
Fig 2.
Time-course of biomass formation (A), glucose consumption (B), pH-changes (C), OPC-4-formation (D), JA-formation (E) and 11/12-hydroxy-JA formation (F) by L. theobromae strain 2334. Cultures were grown in 100 mL (blue line), 250 mL (red line) and 500 mL (green line) flasks with KNO3 as nitrogen source. Additionally, cultures were cultivated in 250 mL flasks containing medium supplemented with NH4NO3 as nitrogen source (violet line). Data are mean of 2–4 replicates. Two replicates: Red– 1.5 d, green– 1.5/3/6 d, violet 1.5 d and the glucose consumption. Four replicates: Red/green/violet– 12 d. The other data represent three replicates. Abbreviations: JA, jasmonic acid; OPC-4, 3-oxo-2-(pent-2-enyl)-cyclopentane-1-butanoic acid.
When KNO3 was used, the fungal cultures reached the stationary phase after 6 days (d) and decreased at the following time-points (Fig 2A). The biomass-yield, basing on glucose as substrate (Yx/s, g biomass/g glucose) gave similar results for the different culture volumes: 100 mL, Yx/s = 0.32; 250 mL, Yx/s = 0.31; 500 mL, Yx/s = 0.35. This suggests that fungal growth was independent from the culture volume. However, when NH4NO3 was used as nitrogen source the fungal biomass-yield was reduced by a factor of 2 (Yx/s = 0.15).
Temporal changes of the glucose uptake showed an inverse trend as the biomass accumulation (Fig 2B). When cultures were grown in the presence of KNO3 or NH4NO3, glucose was completely consumed within 6 d and 9 d, respectively. Within the same time window of day 1 to day 6, we observed drastic pH-changes that were dependent on the nitrogen source: whereas the pH immediately dropped to pH 3, when the fungus was grown in medium supplemented with NH4NO3, a pH increase to pH 8 was observed when KNO3 was supplied (Fig 2C).
Next, we were interested in the metabolic consequences of the different cultivation conditions with regard to JA, its potential precursors OPDA and OPC-4, as well as its potential catabolic derivative 11/12-hydroxy-JA. For this purpose, we analysed the culture filtrate at different time-points of cultivation [30]. We not only detected formation of OPC-4 and 11/12-hydroxy-JA but also found trace amounts of OPDA. However, this compound was measured in very low concentrations (< 0.01 mg/L) and could only be detected after 3 d of cultivation. At the same time point, OPC-4 started to accumulate (Fig 2D). From day 6 to day 8, OPC-4-formation was maximal and declined during the following time points (day 9—day 12) when KNO3 was used as nitrogen source. The amount of OPC-4 that was formed over the whole time course was similar for cultures grown in 100 mL and 500 mL flasks. However, when grown under identical conditions but in 250 mL flasks, the amount of OPC-4 was increased by a factor of 2–3. When NH4NO3 was supplied as a nitrogen source, no OPC-4 production could be observed. JA accumulation started in cultures supplemented with KNO3 approximately at day 3 (Fig 2E). Here, we interestingly observed that the culture size directly affected the amount of JA been synthesized. In cultures grown in 100 mL and 250 mL flasks, JA accumulated after 6–12 d to approx. 420 mg/L and 550 mg/L, respectively, which corresponds to a final product yield of Yp/s = 0.01 mg JA/g glucose. However, when grown in 500 mL flasks L. theobromae strain 2334 formed up to 1250 mg/L JA corresponding to a product yield of Yp/s = 0.03 mg JA/g glucose. For cultures grown in medium supplemented with NH4NO3 formation of JA was highly reduced, yielding only levels of 0.17 – 0.4 mg/L which corresponds to a product yield of Yp/s = 10 ng JA/g glucose. Beside the formation of JA and its potential precursors OPC-4 and OPDA, we also observed formation of the oxidized JA-derivative(s) 11/12-hydroxy-JA. In the plant field, a growing body of evidence suggests that this compound is part of the JA catabolic pathway [31, 33, 37].
Formation of 11/12-hydoxy-JA was also observed to start approx. at 3 d (Fig 2F). Here, we found that the culture volume directly influenced the maximal amount of 11/12-hydroxy-JA that had been formed. Up to 200 mg/L of 11/12-hydroxy-JA was formed by L. theobromae strain 2334 when cultivated in 500 mL flasks with KNO3 as nitrogen source. In smaller flasks (i.e. 100 mL and 250 mL) the amount of this metabolite was reduced by a factor of 1.5–2.
Quantification of plant secondary metabolites produced by L. theobromae strain 2334 under different cultivation conditions
Next we measured plant hormones in extracts obtained from the culture filtrate. We detected very high amounts of indole-3-carboxylic acid (ICA) and small amounts of salicylic acid (SA) [30](Table 1). When KNO3 was used as nitrogen source, production of both compounds increased with the culture volume. Here, the concentration of ICA in the 500 mL culture after 9 days was with 2825 mg/L even higher than the respective amount of JA (Fig 2E). The amounts of SA were much lower but still increasing with the culture volume. In the presence of NH4NO3 the amounts of ICA and SA were strongly reduced. Conclusively, production of ICA as well as SA followed a similar trend as JA-production and increased when the culture size was increased.
Table 1. Average concentrations of plant secondary metabolites, indole-3-carboxylic acid (ICA) and salicylic acid (SA) in the culture medium of L. theobromae strain 2334.
Culture filtrate was obtained after 9 days of growth in medium containing KNO3 as nitrogen source and cultivated as surface culture of different volumes: 100 mL, 250 mL and 500 mL. An additional experiment was performed as surface culture of 250 mL using medium with NH4NO3 as nitrogen source (250 ml-NH4NO3). Data are means ± standard deviations of three replicates. Detection limit for SA was 1x10-4 mg/L at a signal to noise ratio of three.
| Yield (mg/L) | ||||
|---|---|---|---|---|
| Phytohormone | 100 ml | 250 ml | 500 ml | 250 ml-NH4NO3 |
| ICA | 269.25 ± 129.62 | 347.38 ± 78.20 | 2825.32 ± 276.11 | 0.19 ± 0.21 |
| SA | 0.81 ± 0.07 | 1.08 ± 0.30 | 4.67± 5.02 | 0.75 x 10−3± 5 x 10−5 |
Modelling of cellular growth and JA production reveals JA as a fungal secondary metabolite
An attempt of modelling the cellular growth was also carried out by fitting the experimental data to a logistic model (Eq 1) where X is the biomass (g/L) at a specific moment of the cultivation time t (h), Xo and Xmax are the initial and maximum biomass concentration and μm the maximum specific growth rate (h-1). The experimental data for cultures grown in KNO3 containing medium described above were fitted with the logistic model through STATGRAPHIC Centurion version 15.1.0.2 (Stat Point Inc.) that uses the Marquardt-Levenberg algorithm to determine the parameters.
| (1) |
The obtained values defining the model are presented in Table 2 for the different culture volumes. These data illustrate that fungal growth was not significantly affected by the different culture volumes. In a similar approach, we next modelled the time-dependent formation of JA using a logistic model (Eq 2), in which P is the JA production at a specific moment of cultivation time (t), Po and Pmax the initial and maximum JA production, and μp the maximum relative JA production rate.
Table 2. Parameters defining the logistic model of fungal growth.
L. theobromae strain 2334 was cultivated in shake flasks of 100 mL, 250 mL and 500 mL in the presence of KNO3 as nitrogen source.
| Growth Parameters | 100 ml | 250 ml | 500 ml |
|---|---|---|---|
| Xo (g/L) | 0.70 | 0.74 | 0.78 |
| Xmax (g/L) | 16.1 | 18.6 | 17.5 |
| μm (h-1) | 0.04 | 0.04 | 0.04 |
| Regression Coefficient R2 | 0.972 | 0.982 | 0.981 |
| (2) |
The values of the equation parameters are shown in Table 3: While the maximal relative production rate μp observed for the different cultivation conditions was highly similar (0.07–0.06 h-1). The maximum of JA production increased with increased culture volume.
Table 3. Parameters defining the logistic model of JA-production.
L. theobromae strain 2334 was cultivated in shake flasks of 100 mL, 250 mL and 500 mL in the presence of KNO3 as nitrogen source.
| JA production parameters | 100 ml | 250 ml | 500 ml |
|---|---|---|---|
| Po (mg/L) | 0.11 | 0.15 | 0.11 |
| Pmax (mg/L) | 420 | 768 | 1357 |
| μP (h-1) | 0.07 | 0.06 | 0.06 |
| Regression Coefficient R2 | 0.991 | 0.997 | 0.997 |
The correlation between growth and metabolite production can be portrayed by the Luedeking and Piret model [38] and it is represented by the following equation:
| (3) |
with rX and rP being the biomass and metabolite production rate, X the biomass concentration and m and n the parameters of the model. The model can also be expressed in an integrated form [39], as result of combining Eqs 1 and 3 as follows:
| (4) |
Xmax, Xo, μm, P and Po are the parameters defined in the logistic model, m and n the growth and non-growth associated parameters. According of the values of m and n the products of the process are classified as growth associated (m > 0 and n = 0), non-growth associated (m = 0, n > 0) or mixed growth associated (m > 0 and n > 0). The parameters m and n of the model were determined this way and are summarized in Table 4. The obtained data demonstrate that JA-production is a non-growth associated process. This finding thus clearly suggests that JA produced by L. theobromae strain 2334 is a typical fungal secondary metabolite.
Table 4. Parameters defining the Leudiking and Piret model in shake flasks of L. theobromae strain 2334 of volumes of 100 mL, 250 mL and 500 mL and in the presence of KNO3 as nitrogen source.
| Parameters of logistic model | 100 ml | 250 ml | 500 ml |
|---|---|---|---|
| Po (mg/L) | 0 | 0 | 0 |
| m (mg/g biomass) | 0 | 0 | 0 |
| n (mg/g biomass/h) | 3.70 | 3.15 | 5.79 |
| Regression Coefficient R2 | 0.966 | 0.989 | 0.980 |
The integrated form of the modified form of the Luedeking-Piret equation [40] was used to describe the substrate consumption over time (Eq 5).
| (5) |
where α and β, are the Luedeking-Piret equation parameters for substrate consumption.
As shown in Table 5 we found high regression coefficients R2 that indicate a good agreement of the experimental data obtained and the calculated functions. Thus, we are confident that the use of the logistic model to describe the fermentation-process of L. theobromae strain 2334 with regard to biomass-production, JA-formation, and substrate consumption was adequate.
Table 5. Substrate parameters defining of the Leudiking and Piret model in shake cultures of L. theobromae strain 2334 in volumes of 100, 250 and 500 mL.
| Substrate parameters | 100 ml | 250 ml | 500 ml |
|---|---|---|---|
| So (mg/L) | 60.8 | 57.8 | 58.9 |
| α (gS/L/g biomass) | 3.80 | 2.94 | 2.20 |
| β (gS/L/g biomass/h) | 2.54 x 10−4 | 1.53 x 10−3 | 4.02 x 10−3 |
| Regression Coefficient R2 | 0.980 | 0.999 | 0.999 |
Identification of main secondary metabolites of L. theobromae strain 2334 culture filtrate
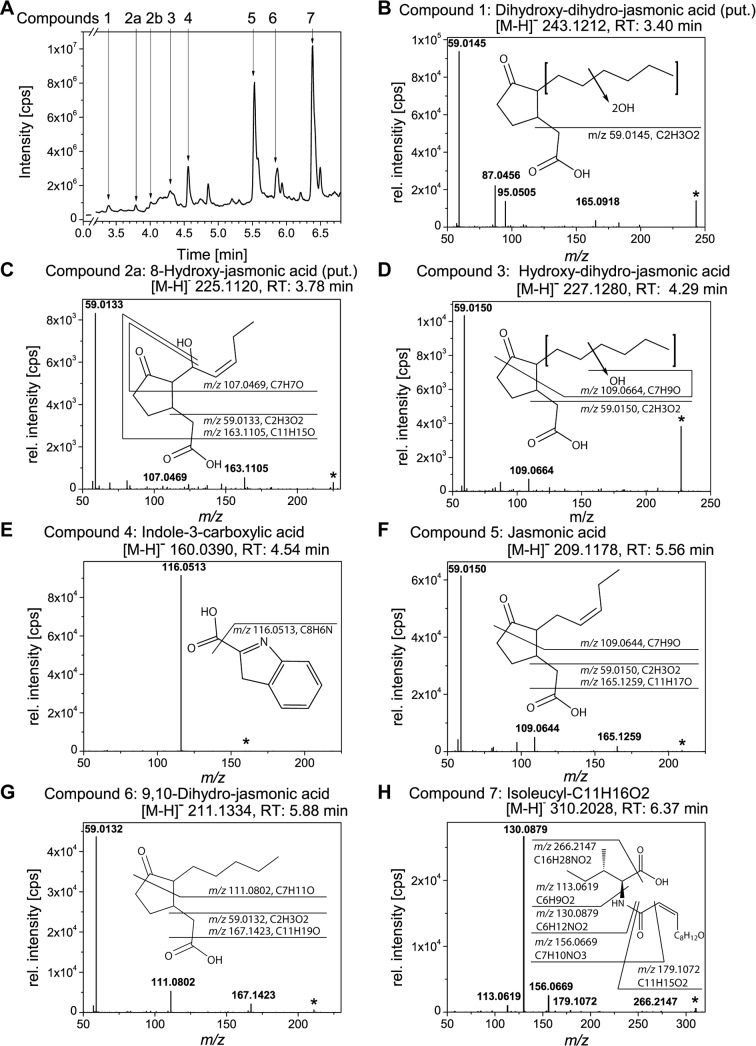
We were interested in the identification of further secondary metabolites that were produced by L. theobromae strain 2334 under conditions optimal for JA-biosynthesis. After extraction from the medium, we analyzed the different metabolites by using UHPLC-ESI-QTOF MS. A total ion chromatogram representative for the MS only mode of the UHPLC-ESI-QTOF-MS analysis is presented in Fig 3A. The majority of the compounds eluted from 3.4 min to 7.0 min. On the basis of the accurate mass and the MS/MS fragmentation patterns, we tentatively assigned the chemical structure of seven of those metabolites (compound 1–7, Fig 3). Compound 1 eluted at 3.40 min and exhibited an accurate mass signal of [M-H]- 243.121 (C12H19O5-, Fig 3B). The MS/MS-spectrum of this compound showed the base-peak at m/z 59.0145. Representing the diagnostic fragment of the carboxy-methyl group from the JA-backbone, this suggests that compound 1 is a JA-derivative [36]. The signal at m/z 165.0918 (C10H13O2) is assumed to result from the loss the carboxy-methyl group as well as the loss of water from one of the proposed hydroxyl residues. In addition two fragment ions of m/z 87.0456 and m/z 95.0505 were detected, which represent fragments of the sum formulas of C4H7O2- and C6H7O-, respectively. We tentatively assign this compound as a dihydroxy-9,10-dihydro-JA. However, the analysis did not allow to determine the exact position of both hydroxy-groups.
Fig 3. UHPLC-ESI-QTOF-MS analysis of the main metabolites in the culture medium of L. theobromae strain 2334.
L. theobromae strain 2334 was cultivated in medium supplemented with KNO3 for 9 d. After extraction, compounds of the culture filtrate were analyzed via UHPLC-ESI-QTOF-MS. Shown is a total ion chromatogram (A), as well as high resolution MS/MS-spectra (negative ionization mode; collision energy 10 eV) of different compounds (B-H). Proposed structures and fragmentations are shown as inset for each spectrum. The following compounds were identified: dihydroxy-dihydro-JA ([M-H]- 243.1212, RT 3.40 min) (B), 8-hydroxy-JA ([M-H]- 225.1120, RT 3.78 min) (C), hydroxy-dihydro-JA ([M-H]- 227.1280, RT 4.29 min) (D), Indol-3-carboxylic acid ([M-H]- 160.0390, RT 4.54 min) (E), JA ([M-H]- 209.1178, RT 5.56 min) (F), 9,10-dihydro-JA ([M-H]- 211.1334, RT 5.88 min) (G), Isoleucyl-C11H16O2. ([M-H]- 310.2028, RT 6.37 min) (H). Abbreviations: JA, jasmonic acid.
The following two compounds eluted at 3.78 min and at 4.02 min, respectively (Fig 3A, compound 2a and 2b). Both exhibited an identical accurate mass of [M-H]- 225.1120 and gave similar MS/MS-spectra (compound 2a (Fig 3C) and 2b (Fig 1B)). Comparison of the retention times with that of authentic standards suggested that compound 2b is either 11- or 12-hydroxy-JA or a mixture of both (11/12-hydroxy-JA, see above). The chemical structure of compound 2a could not be fully resolved with respect to the position of the hydroxy-group. However, the shorter retention time suggests that the hydroxy group is located at the C8 as indicated in the inset of Fig 3C. 8-hydroxy-JA was described in the supernatant of L. theobromae by Miersch and co-workers [19].
The MS/MS-analysis of compound 3 is shown in Fig 3D. This compound eluted at 4.29 min with an accurate mass signal of [M-H]- 227.1280. The MS/MS-spectrum resembled that of compound 2a and 2b. Again, we found the base-peak at m/z 59.0150 (carboxy-methyl group) suggesting that this compound is also a JA-derivative. The fragment ion at m/z 109.0664 (corresponding to the sum formula of C7H9O-) is shifted by the mass of two hydrogens to higher m/z-ratios compared to that of hydroxy-JAs (Fig 1B) and (Fig 3C). Those findings allowed the tentative assignment of compound 3 as a hydroxy-dihydro-JA.
Compound 4 eluted at 4.54 min and exhibited an accurate mass signal of [M-H]- 160.0390. The MS/MS-spectrum (Fig 3E) displayed a highly intense base peak at m/z 116.0513 ([M-CO2-H]-, neutral loss of the carboxyl-group as CO2) and a very weak signal of the parent-ion at m/z 160.0390 [M-H]-. Comparison of the fragmentation pattern of compound 4 with that for ICA (Scripps Center for Metabolomics, METLIN data base: MID3795) confirmed the occurrence of ICA in the supernatant of L. theobromae.
Compound 5 (RT 5.56 min and [M-H]- 209.1178) was identified as JA (Fig 3F). In accordance with the spectra of compound 1, 2, and 3, the base peak was again at m/z 59.0150 and additional signals were found at m/z 109.0644 and m/z 165.1259. The latter one had been described as JA-fragment of minor intensity (Scripps Center for Metabolomics, METLIN data base: MID 3345) [41]. In addition, the identity of JA was confirmed by comparing the retention time with that of an authentic standard.
MS/MS-spectrum of compound 6 (RT 5.88 min, [M-H]- 211.1334, Fig 3G) was similar to that of JA in respect to the base-peak at m/z 59.0132. The minor signals of m/z 167.1423 and m/z 111.0802 were shifted to higher m/z-values by the mass of two hydrogens in comparison to the fragments of JA. Based on this result we assigned this compound as dihydro-JA.
Compound 7 eluted at 6.37 min and exhibited an accurate mass signal of [M-H]- 310.2028 (Fig 3H). In order to elucidate the chemical structure of this compound, we performed MS/MS-experiments in the negative and positive ionization mode. However, both analyses together did allow the unequivocal identification of this metabolite. The MS/MS-spectrum of the negatively ionized analyte displayed a base peak at m/z 130.0879, which is characteristic for an isoleucine (Ile) or leucine (Leu) moiety. The signals at m/z 113.0619 (Ile/Leu moiety with a neutral loss of the amino group) as well as m/z 179.1072 ([M-Ile/Leu-H]-) confirmed the substructure. The fragment of m/z 156.0669 allowed to foretell the connection of a fatty acid like moiety (C7H10NO3-) via an amide (peptide) bound to the amino group of the Ile residue. By pseudo MS3 of m/z 132.1020 (Ile/Leu moiety) in the positive mode, Ile could be unequivocally identified as amino acid moiety of compound 7 (presence of the diagnostic Ile-fragment of m/z 41.0388, data not shown). In the positive MS/MS mode compound 7 showed a base-peak at m/z 266.2102 and additional signals at m/z 86.0966, m/z 125.2102 and m/z 198.1467 (data not shown). From all obtained data, we propose a chemical structure of this compound as shown in Fig 3H. The structure comprises of a C11H15O2 unit that is bound to the α-amino-group of an Ile-moiety via an amide bond. Due to an incomplete fragmentation of the proposed fatty acid like structure (C11H15O2 unit) by ESI-MS/MS we could not obtain further structural information.
Discussion
In the present study, we aimed to improve the capacity of L. theobromae strain 2334 to produce JA, JA-derivatives and further secondary metabolites. Here, the focus laid primarily on the analysis of the influence of the nitrogen-source (KNO3 and NH4NO3) and the culture-volume. Only in medium containing KNO3, formation of JA was observed indicating that the choice of the nitrogen source is an important parameter for JA biosynthesis. Our results further demonstrate that JA production is highly dependent on the culture volume. We observed that the use of 500 mL flasks for cultivation led to the formation of up to 1.25 g/L JA after 9 days. This may be explained by an optimal oxygen and nutrient diffusion within the culture medium under these conditions. A similar trend has previously been reported by Dhandhukia and co-workers who obtained up to 170 mg/L JA when L. theobromae strain MTCC-3068 was grown in 1 L flasks [14]. This amount is reduced by a factor of 7–8 compared to JA formed by the L. theobromae strain used in this study. One explanation for this significant difference might be the varying inherent capacities of the two L. theobromae strains to synthesize JA. On the other hand, Dhandhukia and co-workers used NaNO3 as nitrogen source [14] whereas we used KNO3 for cultivation. As discussed above and also reported elsewhere the choice of nutritional parameters might directly influence the amount of JA produced by L. theobromae [15].
To deepen our understanding on JA-production by L. theobromae and thus to possibly further enhance the amount of JA that is formed, we additionally investigated the anabolism and catabolism of JA. For this purpose we analyzed the temporal changes in the amount of JA and also examined the cultures for metabolites that might be formed upstream (e.g. OPDA and OPC-4) or downstream of JA (hydroxy-JA derivatives) and monitored temporal alterations in their production. Interestingly, we detected the formation of OPDA at day 3 albeit in very minor concentrations (data not shown). Free OPDA has not been detected in L. theobromae before. In 2010, however, Tsukada and co-workers observed the transient production of the methyl-ester of OPDA [42]. From this finding it was hypothesized that L. theobromae might synthesize JA via a pathway analogous to that from plants [42]. In order to evaluate this idea, we further analyzed the cultivation medium and detected at defined time-points of growth significant amounts of OPC-4 (Fig 1A and Fig 2D). In plants, this metabolite presents the direct precursor of JA that is formed from OPDA after reduction and two-rounds of β-oxidation. Using the cultivation conditions described above, L. theobromae strain 2334 formed OPC-4 from day 3 to day 12 (Fig 2D). Within the same time-window, JA-accumulation was observed (Fig 2E). This strongly suggests for a metabolic connection of these metabolites in L. theobromae and our results showed that the volume of the culture influences the rate of OPC-4 conversion into JA.
Beside the formation of anabolites of JA, like OPC-4, we as analyzed potential catabolites of JA. In plants, ω-oxidation of its pentenyl-side chain is discussed to be one possible way to inactivate JA-signaling [31–34, 37, 43–45]. In order to further evaluate JA-catabolism and to examine potential parallels between plants and fungal JA-metabolism, we analyzed fungal cultures at different time-points for hydroxy-JA derivatives. Interestingly, we could detect formation of 11/12-hydroxy-JA starting at day 3 (Fig 2F). Based on these findings, we hypothesized that fungal and plant JA-metabolism may involve similar reaction steps.
Several studies reported on the capability of fungal species to produce JA [14, 16, 18, 20, 27, 42, 46]. However, only recently the biotechnological capacity to produce this metabolite has been recognized and attempts to systematically optimize the cultivation conditions were reported [14–16]. In this study, we attempted to model fungal growth and to relate this parameter to JA-production and substrate/glucose consumption. By this analysis, we on the one hand gathered information on parameters of JA-production and fungal growth that may be important for the development and further optimization of fungal cultivation. On the other hand, our results also demonstrate that JA-production is a non-growth related process and thus corroborate that JA is a compound of the secondary metabolism in the fungus.
As mentioned above JA plays a key role in plant development and plant defense and thereby also orchestrates the metabolism of different phytohormones. In order to manipulate those stress- and defense reactions of infected plants on a metabolic level, several fungal species adopted the capacity to produce phytohormones and further plant secondary metabolites (for recent reviews on this topic see [47, 48]). To further test this idea and thereby to gather more information on fungal JA-metabolism, we analyzed whether additional phytohormones and secondary metabolites are produced by L. theobromae beside JA. Our results demonstrate that L. theobromae strain 2334 synthesizes several plant related compounds of the secondary metabolism, which were detectable in the culture filtrate. Interestingly, one of those metabolites was identified as chorismate (data not shown). This compound may be regarded as the starting point for tryptophan-biosynthesis and is thus an important constituent of the so-called tryptophan-derived secondary metabolism [30] and also of indole acetic acid (IAA) biosynthesis in plants [49]. In addition, chorismate serves as precursor of salicylic acid, a compound that plays a key-role in the regulation of different physiological processes [50]. Besides minor amounts of chorismate, we found that trace amounts of IAA (0.03–0.07 mg.L-1, data not shown) were formed by L. theobromae strain 2334. Although IAA has been mainly detected in plants, also microorganisms as well as some animals have the capacity to produce IAA (i.e. [51]). In plants, IAA functions as regulator of plant development, whereas in animals antitumor activity has been reported. In addition, antimicrobial activity was observed for some IAA-derivatives [13].
Another metabolite that plays a role in the tryptophan-derived secondary metabolism, which was detected in high amounts in the fungal filtrate, was ICA. This indolic metabolite is formed upon pathogen infection in plants [52] and has also been detected in several microorganisms [53]. It is interesting to note, that two different routes for the biosynthesis of ICA have been proposed for plants and microorganism. While microbial ICA-formation is thought to involve IAA as precursor, plants use a different route that is independent from IAA [52, 53]. Recently, formation of ICA was also observed in L. theobromae strain ME4-2 that was isolated from floral parts of mistletoe. Since only minor amounts of IAA (0.03–0.07 mg.L-1) could be detected in this study, it was proposed that ICA in this L. theobromae species is formed via an IAA-independent route [54]. This finding may thus suggest an ICA-biosynthetic route that is common for microorganisms as proposed by [53]. In line with the detection of ICA in L. theobromae strain 2334 is a recent study by Castillo and co-workers who detected indole-3-butyric acid and indole-3-propionic acid—two further indole-derivatives—beside IAA. However, formation of ICA was not reported in that work [21].
About thirty years ago Miersch and co-workers observed formation of (+)-7-iso JA, (+)-9,10-dihydro-7-iso JA, (+)-11,12-didehydro-7-iso JA, and (+)-cucurbic acid as well as a large variety of hydroxylated JA-derivatives including 8-, 11- and 12-hydroxy-JA in the culture filtrate of L. theobromae strain D 7/2. Later, further JA-derived molecules were identified and termed lasiojasmonates [46, 55] suggesting that a large variety of JA-derived metabolites are formed by L. theobromae. In order to expand the knowledge on the repertoire of the different jasmonates formed by L. theobromae-species, we analyzed the main secondary metabolites secreted into the medium. Besides the already reported structures of JA, 9,10-dihydro-JA and 11/12-hydroxy-JA, we identified two further oxidized dihydro-JA-derivatives: hydroxy-9,10-dihydro-JA and dihydroxy-9,10-dihydro-JA. Formation of the analogous 11-hydroxy derivative in Aspergillus niger has been reported before [23]. A further oxidized JA-derivative (compound 2a) was detected that gave the same MS/MS-spectrum as 11/12-hydoxy-JA but eluted at an earlier retention-time suggesting an enhanced polarity of this compound. Potential positions for the hydroxyl-group are the C8, C9 and C10 of the JA backbone. Location of the hydroxyl-group at the latter two positions would result in the formation of an enol moiety that might tautomerize to the corresponding keto form via keto-/enol tautomerism. As there is no evidence for a functional keto-group in our analysis and as we anticipate that the resulting keto-tautomere may exhibit a rather increased retention time compared to the hydroxy-isomers, we tentatively assign compound 2a as 8-hydroxy-JA. This idea is supported by the above mentioned study of Miersch and co-workers that showed the formation of 8-hydroxy-JA by L. theobromae [19]. Unfortunately, no authentic standard was available to test our hypothesis.
An oxygenase that has the capacity to oxidize JA has been identified recently in fungi [56] but remains unidentified in plants. Here, ω-oxidation has only been reported to occur at the respective Ile conjugate—JA-Ile—and is considered as one possibility to inactivate the JA-signaling activity [31, 43, 45]. Formation of JA-Ile and further JA-amino acid conjugates by L. theobromae strain 2334 (JA-Gly, JA-Ser, JA-Thr) has been reported recently [21]. We did not detect any JA-amino acid conjugate produced by the L. theobromae-strain used in this study. Castillo and co-workers described the signal of m/z 310.2028 in the supernatant of L. theobromae strain 2334 as JA-Tyr [21]. We obtained similar mass and fragmentation pattern for compound 7, but the high resolution mass information of our analysis rebut the identity as JA-Tyr. Our analysis indicated that compound 7, one of the main metabolites of the fungal culture filtrate, consisted of Ile-residue that was bound to a C11-moiety via an amide-bond and could thus reminiscent to a JA-Ile structure shortened by a CH2-moiety.
Conclusion
In this study, we used the fungus L. theobromae to produce JA and optimized the cultivation parameters to enhance the final JA-yield. Using those parameters, we obtained from a single 100 mL fungal culture as much JA as that is produced by more than 1000 Jasminum sambac blossoms. Furthermore, we establish a mathematic model that describes and correlates fungal growth and JA-production and thus may provide a basis for upscaling processes in industrial fermentation. Our results also demonstrate that beside JA further metabolites are formed that have originally been identified in plants. They suggest that fungal JA-metabolism involves similar reactions as in plants. A deeper understanding of the inherent function of JA and its metabolic pathways in fungi will help to further improve JA production and is an interesting topic for further investigations.
Supporting Information
In this table, the raw data of the different experiments are listed.
(XLSX)
Acknowledgments
FE was supported through a Deutscher Akademischer Austauschdienst scholarship (DAAD, 50015559) and IF by the Deutsche Forschungsgemeinschaft (ZUK 45/2010). IF, SH and DR were supported by the IRTG 2172 “PRoTECT” of the Göttingen Graduate School of Neuroscience and Molecular Biology (GGNB). We thank Pia Meyer and Pablo Tarazona for excellent assistance and Dr. Otto Miersch (IPB, Halle/Saale, Germany) for providing jasmonate standards as well as for fruitful discussions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
FE was supported through a Deutscher Akademischer Austauschdienst scholarship (DAAD, 50015559) and IF by the Deutsche Forschungsgemeinschaft (ZUK 45/2010). IF, SH and DR were supported by the IRTG 2172 "PRoTECT" of the Göttingen Graduate School of Neuroscience and Molecular Biology (GGNB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Busch R, Hirth T, Liese A, Nordhoff S, Puls J, Pulz O, et al. The utilization of renewable resources in German industrial production. Biotechnol J. 2006;1: 770–6. 10.1002/biot.200600057 [DOI] [PubMed] [Google Scholar]
- 2.Biermann U, Bornscheuer U, Meier MAR, Metzger JO, Schäfer HJ. Oils and fats as renewable raw materials in chemistry. Angew Chem Int Ed Engl. 2011;50: 3854–71. 10.1002/anie.201002767 [DOI] [PubMed] [Google Scholar]
- 3.Song MC, Kim EJ, Kim E, Rathwell K, Nam SJ, Yoon YJ. Microbial biosynthesis of medicinally important plant secondary metabolites. Nat Prod Rep. 2014;31: 1497–509. 10.1039/c4np00057a [DOI] [PubMed] [Google Scholar]
- 4.Ueda J, Miyamoto K, Aoki M, Hirata T, Sato T, Momotani Y. Identification of jasmonic acid in Chlorella and Spirulina. Bull Univ Osaka Pref Ser B. 1991;43: 103–8. [Google Scholar]
- 5.Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Ann Rev Plant Physiol Plant Mol Biol. 1997;48: 355–81. [DOI] [PubMed] [Google Scholar]
- 6.Miersch O, Günther T, Fritsche W, Sembdner G. Jasmonates from different fungal species. Nat Prod Lett. 1993;2: 293–9. [Google Scholar]
- 7.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111: 1021–58. 10.1093/aob/mct067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaller A, Stintzi A. Enzymes in jasmonate biosynthesis—Structure, function, regulation. Phytochemistry. 2009;70: 1532–8. 10.1016/j.phytochem.2009.07.032 [DOI] [PubMed] [Google Scholar]
- 9.Demole E, Lederer E, Mercier D. Isolement et détermination de la structure du jasmonate de méthyle, constituant odorant caractéristique de l'essence de jasmin. Helv Chim Acta. 1962;45: 675–85. [Google Scholar]
- 10.Scognamiglio J, Jones L, Letizia CS, Api AM. Fragrance material review on methyl jasmonate. Food Chem Toxicol. 2012;50 Suppl 3: 572–6. [DOI] [PubMed] [Google Scholar]
- 11.Ghasemi Pirbalouti A, Sajjadi SE, Parang K. A review (research and patents) on jasmonic acid and its derivatives. Arch Pharm. 2014;347: 229–39. [DOI] [PubMed] [Google Scholar]
- 12.Raviv Z, Cohen S, Reischer-Pelech D. The anti-cancer activities of jasmonates. Cancer Chemother Pharmacol. 2013;71: 275–85. 10.1007/s00280-012-2039-z [DOI] [PubMed] [Google Scholar]
- 13.Lin L, Tan RX. Cross-kingdom actions of phytohormones: a functional scaffold exploration. Chem Rev. 2011;111: 2734–60. 10.1021/cr100061j [DOI] [PubMed] [Google Scholar]
- 14.Dhandhukia PC, Thakkar VR. Standardization of growth and fermentation criteria of Lasiodiplodia theobromae for production of jasmonic acid. Afric J Biotechnol. 2007;6: 707–12. [Google Scholar]
- 15.Dhandhukia PC, Thakkar VR. Response surface methodology to optimize the nutritional parameters for enhanced production of jasmonic acid by Lasiodiplodia theobromae. J Appl Microbiol. 2008;105: 636–43. 10.1111/j.1365-2672.2008.03803.x [DOI] [PubMed] [Google Scholar]
- 16.Eng F, Gutiérrez-Rojas M, Favela-Torres E. Culture conditions for jasmonic acid and biomass production by Botryodiplodia theobromae in submerged fermentation. Proc Biochem. 1998;33: 715–20. [Google Scholar]
- 17.Miersch O, Brückner B, Schmidt J, Sembdner G. Cyclopentane fatty acids from Gibberella fujikuroi. Phytochemistry. 1992;31: 3835–7. [Google Scholar]
- 18.Aldridge DC, Galt S, Giles D, Turner WB. Metabolites of Lasiodiplodia theobromae. J Chem Soc C: Organic. 1971: 1623–7. [Google Scholar]
- 19.Miersch O, Schneider G, Sembdner G. Hydroxylated jasmonic acid and related compounds from Botryodiplodia theobromae. Phytochemistry. 1991;30: 4049–51. [Google Scholar]
- 20.Miersch O, Preiss A, Sembdner G, Schreiber K. (+)-7-iso-jasmonic acid and related compounds from Botryodiplodia theobromae. Phytochemistry. 1987;26: 1037–9. [Google Scholar]
- 21.Castillo G, Torrecillas A, Nogueiras C, Michelena G, Sanchez-Bravo J, Acosta M. Simultaneous quantification of phytohormones in fermentation extracts of Botryodiplodia theobromae by liquid chromatography-electrospray tandem mass spectrometry. World J Microb Biot. 2014;30: 1937–46. [DOI] [PubMed] [Google Scholar]
- 22.Miersch O, Bohlmann H, Wasternack C. Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry. 1999;50: 517–23. [Google Scholar]
- 23.Miersch O, Porzel A, Wasternack C. Microbial conversion of jasmonates—hydroxylations by Aspergillus niger. Phytochemistry. 1999;50: 1147–52. [DOI] [PubMed] [Google Scholar]
- 24.Cross BE, Webster GRB. New metabolites of Gibberella fujikuroi. Part XV. N-jasmonoyl-isoleucine and N-dihydrojasmonoyl-isoleucine. J Chem Soc C: Organic. 1970: 1839–&. [DOI] [PubMed] [Google Scholar]
- 25.Brodhun F, Cristobal-Sarramian A, Zabel S, Newie J, Hamberg M, Feussner I. An iron 13S-lipoxygenase with an α-linolenic acid specific hydroperoxidase activity from Fusarium oxysporum. PLoS ONE. 2013;8: e64919 10.1371/journal.pone.0064919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann I, Oliw EH. Discovery of a linoleate 9S-dioxygenase and an allene oxide synthase in a fusion protein of Fusarium oxysporum. J Lipid Res. 2013;54: 3471–80. 10.1194/jlr.M044347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jernerén F, Eng F, Hamberg M, Oliw E. Linolenate 9R-dioxygenase and allene oxide synthase activities of Lasiodiplodia theobromae. Lipids. 2012;47: 65–73. 10.1007/s11745-011-3622-5 [DOI] [PubMed] [Google Scholar]
- 28.Deeg R, Kraemer W, Ziegenhorn J. Kinetic determination of serum glucose by use of the hexokinase/glucose-6-phosphate dehydrogenase method. J Clin Chem Clin Biochem. 1980;18: 49–52. [DOI] [PubMed] [Google Scholar]
- 29.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49: 1137–46. 10.1194/jlr.D700041-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iven T, König S, Singh S, Braus-Stromeyer SA, Bischoff M, Tietze LF, et al. Transcriptional activation and production of tryptophan-derived secondary metabolites in Arabidopsis roots contributes to the defense against the fungal vascular pathogen Verticillium longisporum. Mol Plant. 2012;5: 1389–402. 10.1093/mp/sss044 [DOI] [PubMed] [Google Scholar]
- 31.Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Desaubry L, et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J Biol Chem. 2012;287: 6296–306. 10.1074/jbc.M111.316364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitaoka N, Kawaide H, Amano N, Matsubara T, Nabeta K, Takahashi K, et al. CYP94B3 activity against jasmonic acid amino acid conjugates and the elucidation of 12-O-β-glucopyranosyl-jasmonoyl-l-isoleucine as an additional metabolite. Phytochemistry. 2014;99: 6–13. 10.1016/j.phytochem.2013.12.019 [DOI] [PubMed] [Google Scholar]
- 33.Koo AJK, Cooke TF, Howe GA. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci USA. 2011;108: 9298–303. 10.1073/pnas.1103542108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widemann E, Heitz T, Miesch L, Miesch M, Heinrich C, Pinot F, et al. Identification of the 12-oxojasmonoyl-isoleucine, a new intermediate of jasmonate metabolism in Arabidopsis, by combining chemical derivatization and LC-MS/MS analysis. Metabolomics. 2015;11: 991–7. [Google Scholar]
- 35.Bao J, Gao X, Jones AD. Unusual negative charge-directed fragmentation: collision-induced dissociation of cyclopentenone oxylipins in negative ion mode. Rapid Commun Mass Spectrom. 2014;28: 457–64. 10.1002/rcm.6803 [DOI] [PubMed] [Google Scholar]
- 36.Durgbanshi A, Arbona V, Pozo O, Miersch O, Sancho JV, Gomez-Cadenas A. Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography-electrospray tandem mass spectrometry. J Agric Food Chem. 2005;53: 8437–42. 10.1021/jf050884b [DOI] [PubMed] [Google Scholar]
- 37.Kitaoka N, Matsubara T, Sato M, Takahashi K, Wakuta S, Kawaide H, et al. Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 2011;52: 1757–65. 10.1093/pcp/pcr110 [DOI] [PubMed] [Google Scholar]
- 38.Luedeking R, Piret EL. A kinetic study of the lactic acid fermentation. Batch process at controlled pH. Reprinted from Journal of Biochemical and Microbiological Technology Engineering Vol. I, No. 4. Pages 393–412 (1959). Biotechnol Bioeng. 2000;67: 636–44. [DOI] [PubMed] [Google Scholar]
- 39.Marques AM, Estanol I, Alsina JM, Fuste C, Simon-Pujol D, Guinea J, et al. Production and rheological properties of the extracellular polysaccharide synthesized by Pseudomonas sp. Strain EPS-5028. Appl Environ Microbiol. 1986;52: 1221–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss RM, Ollis DF. Extracellular microbial polysaccharides. I. Substrate, biomass, and product kinetic equations for batch xanthan gum fermentation. Biotechnol Bioeng. 1980;22: 859–73. [Google Scholar]
- 41.Segarra G, Jauregui O, Casanova E, Trillas I. Simultaneous quantitative LC-ESI-MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry. 2006;67: 395–401. 10.1016/j.phytochem.2005.11.017 [DOI] [PubMed] [Google Scholar]
- 42.Tsukada K, Takahashi K, Nabeta K. Biosynthesis of jasmonic acid in a plant pathogenic fungus, Lasiodiplodia theobromae. Phytochemistry. 2010;71: 2019–23. 10.1016/j.phytochem.2010.09.013 [DOI] [PubMed] [Google Scholar]
- 43.Koo AJ, Howe GA. Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front Plant Sci. 2012;3: 19 10.3389/fpls.2012.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poudel AN, Zhang T, Kwasniewski M, Nakabayashi R, Saito K, Koo AJ. Mutations in jasmonoyl-L-isoleucine-12-hydroxylases suppress multiple JA-dependent wound responses in Arabidopsis thaliana. Biochim Biophys Acta. 2016. [DOI] [PubMed] [Google Scholar]
- 45.Widemann E, Grausem B, Renault H, Pineau E, Heinrich C, Lugan R, et al. Sequential oxidation of jasmonoyl-phenylalanine and jasmonoyl-isoleucine by multiple cytochrome P450 of the CYP94 family through newly identified aldehyde intermediates. Phytochemistry. 2015;117: 388–99. 10.1016/j.phytochem.2015.06.027 [DOI] [PubMed] [Google Scholar]
- 46.Andolfi A, Maddau L, Cimmino A, Linaldeddu BT, Basso S, Deidda A, et al. Lasiojasmonates A-C, three jasmonic acid esters produced by Lasiodiplodia sp., a grapevine pathogen. Phytochemistry. 2014;103: 145–53. 10.1016/j.phytochem.2014.03.016 [DOI] [PubMed] [Google Scholar]
- 47.Christensen SA, Kolomiets MV. The lipid language of plant-fungal interactions. Fungal Genet Biol. 2011;48: 4–14. 10.1016/j.fgb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 48.Brodhun F, Feussner I. Oxylipins in fungi. FEBS J. 2011;278: 1047–63. 10.1111/j.1742-4658.2011.08027.x [DOI] [PubMed] [Google Scholar]
- 49.Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63: 2853–72. 10.1093/jxb/ers091 [DOI] [PubMed] [Google Scholar]
- 50.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414: 562–5. 10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- 51.Sirrenberg A, Göbel C, Grond S, Czempinski N, Ratzinger A, Karlovsky P, et al. Piriformospora indica affects plant growth by auxin production. Physiol Plant. 2007;131: 581–9. 10.1111/j.1399-3054.2007.00983.x [DOI] [PubMed] [Google Scholar]
- 52.Böttcher C, Chapman A, Fellermeier F, Choudhary M, Scheel D, Glawischnig E. The biosynthetic pathway of indole-3-carbaldehyde and indole-3-carboxylic acid derivatives in Arabidopsis. Plant Physiol. 2014;165: 841–53. 10.1104/pp.114.235630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis PJ, Gustafson ME, Rosazza JP. Formation of indole-3-carboxylic acid by Chromobacterium violaceum. J Bacteriol. 1976;126: 544–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian CD, Fu YH, Jiang FS, Xu ZH, Cheng DQ, Ding B, et al. Lasiodiplodia sp. ME4-2, an endophytic fungus from the floral parts of Viscum coloratum, produces indole-3-carboxylic acid and other aromatic metabolites. BMC Microbiol. 2014;14: 297 10.1186/s12866-014-0297-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andolfi A, Basso S, Giambra S, Conigliaro G, Piccolo SL, Alves A, et al. Lasiolactols A and B produced by the grapevine fungal pathogen Lasiodiplodia mediterranea. Chem & Biodiv. 2016;13: 395–402. [DOI] [PubMed] [Google Scholar]
- 56.Patkar RN, Benke PI, Qu Z, Constance Chen YY, Yang F, Swarup S, et al. A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nature Chem Biol. 2015;11: 733–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In this table, the raw data of the different experiments are listed.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.